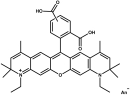
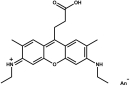
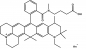
ATTO 647N




|
Optische Eigenschaften |
||||
|
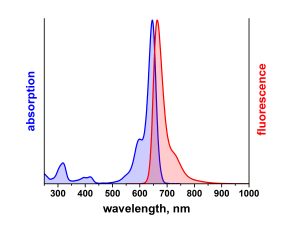
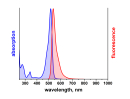
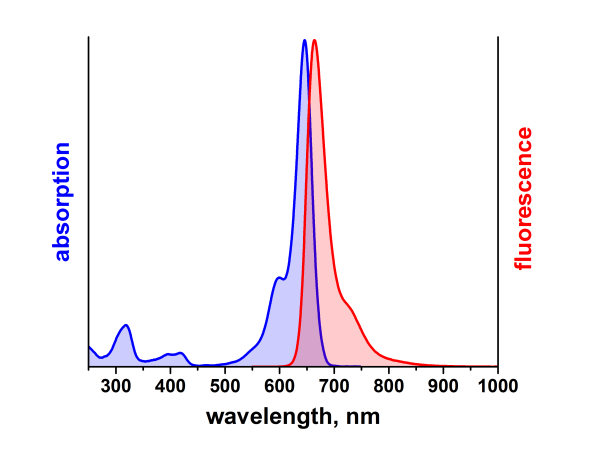
λabs |
646 |
nm |
||
|
εmax |
1,5×105 |
M-1 cm-1 |
||
|
λfl |
664 |
nm |
||
|
ηfl |
65 |
% |
||
|
τfl |
3,5 |
ns |
||
|
CF260 = ε260/εmax |
0,04 |
|
||
|
CF280 = ε280/εmax |
0,03 |
|
||
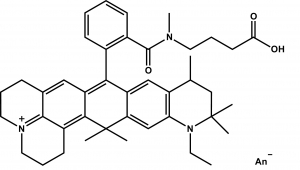
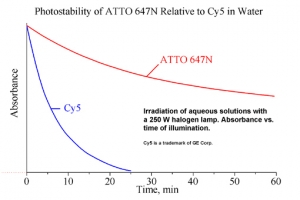
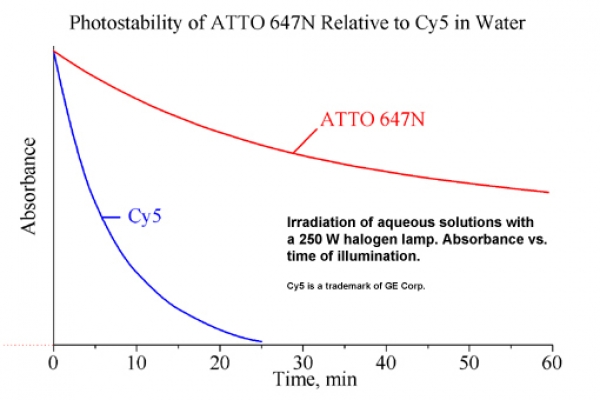
ATTO 647N zählt im roten Spektralbereich zu einer neuen – speziell von ATTO-TEC entwickelten – Generation von Fluoreszenzmarkern. Zu den charakteristischen Eigenschaften des Farbstoffs zählen starke Absorption, sehr hohe Fluoreszenzquantenausbeute, hohe thermische und photochemische Stabilität sowie außergewöhnlich hohe Stabilität gegenüber atmosphärischem Ozon. ATTO 647N ist somit besonders geeignet im Bereich der Einzelmoleküldetektion sowie zur hochauflösenden Mikroskopie (SIM, STED, etc.).
Absorption und Fluoreszenz sind im pH-Bereich von 2 - 11 pH-unabhängig. Der Farbstoff ist mäßig hydrophil. ATTO 647N ist ein kationischer Farbstoff. Nach der Kopplung an ein Substrat ist der Farbstoff einfach positiv geladen. ATTO 647N ist ein Gemisch aus zwei Isomeren mit praktisch identischen Absorptions- und Fluoreszenzeigenschaften. Eine effiziente Anregung der Fluoreszenz erfolgt im Bereich 625 - 660 nm. Für den Farbstoff geeignete Anregungslichtquellen sind neben dem HeNe-Laser (633 nm) der Krypton-Ionen-Laser (647 nm) oder gängige Diodenlaser mit Emission bei 650 nm.
Produktdatenblätter
- ATTO 647N Produktinformation
- NHS-Ester
- Maleimid
- Iodacetamid
- Phalloidin
- Amin
- Azid/Alkin
- Streptavidin
- Tetrazin (MeTet)
Absorptions- und Emissionsspektrum (ASCII)
- ATTO 647N Absorption (.txt)
- ATTO 647N Emission (.txt)
Absorptions- und Emissionsspektrum (Grafik)
- ATTO 647N Absorption/Fluoreszenz (.jpg)
Sicherheitsdatenblätter
- ATTO 647N Carboxy MSDS
- ATTO 647N NHS-Ester MSDS
- ATTO 647N Maleimid MSDS
- ATTO 647N Streptavidin MSDS
- ATTO 647N Biotin MSDS
- ATTO 647N Phalloidin MSDS
- ATTO 647N Amin MSDS
- ATTO 647N Azid MSDS
- ATTO 647N Iodacetamid MSDS
- ATTO 647N Hydrazid MSDS
- ATTO 647N Alkin MSDS
- ATTO 647N Cadaverin MSDS
- ATTO 647N MeTet MSDS
- ATTO 647N Peg(4)-DBCO MSDS
- ATTO 647N Phosphoramidit MSDS
Ausgewählte Literatur zu ATTO 647N:
M. Aleksanyan, A. Grafmüller, F. Crea, V. Georgiev, N. Yandrapalli, S. Block, J. Heberle, R. Dimova, Photomanipulation of Minimal Synthetic Cells, Advanced Science 10, 2304336 (2023).
S. Asfia, R. Seemann, J.-B. Fleury, Phospholipids diffusion on the surface of model lipid droplets, Biochimica et Biophysica Acta (BBA) - Biomembranes 1865, 184074 (2023).
T. Bauer, I. Alberg, L. Zengerling, P. Besenius, K. Koynov, B. Slütter, R. Zentel, I. Que, H. Zhang, M. Barz, Tuning the Cross-Linking Density and Cross-Linker in Core Cross-Linked Polymeric Micelles and Its Effects on the Particle Stability in Human Blood Plasma and Mice, Biomacromolecules 24, 3545 (2023).
M. Bertolini, M. Wong, L. Mendive-Tapia, M. Vendrell, Smart probes for optical imaging of T cells and screening of anti-cancer immunotherapies, Chemical Society reviews 52, 5352 (2023).
R. Cai, Z. Kolabas, C. Pan, H. Mai, S. Zhao, D. Kaltenecker, F. Voigt, M. Molbay, T.-L. Ohn, C. Vincke, M. Todorov, F. Helmchen, J. van Ginderachter, A. Ertürk, Whole-mouse clearing and imaging at the cellular level with vDISCO, Nature Protocols 18, 1197 (2023).
S. Calabrese, A. Markl, M. Neugebauer, S. Krauth, N. Borst, F. von Stetten, M. Lehnert, Reporter emission multiplexing in digital PCRs (REM-dPCRs), The Analyst 9, 143 (2023).
T.-J. Chang, J. Hsu, T. Yang, Single-molecule localization microscopy reveals the ultrastructural constitution of distal appendages in expanded mammalian centrioles, Nature communications 14, 1688 (2023).
A. Chaudhury, S. Swarnakar, G. Pattnaik, G. Varshney, H. Chakraborty, J. Basu, Peptide-Induced Fusion of Dynamic Membrane Nanodomains, Langmuir 39, 17713 (2023).
Z. Cheng, C. Stefani, T. Skillman, A. Klimas, A. Lee, E. DiBernardo, K. Brown, T. Milman, Y. Wang, B. Gallagher, K. Lagree, B. Jena, J. Pulido, S. Filler, A. Mitchell, N. Hiller, A. Lacy-Hulbert, Y. Zhao, MicroMagnify, Advanced Science 10, 2302249 (2023).
E. Di Franco, A. Costantino, E. Cerutti, M. D'Amico, A. Privitera, P. Bianchini, G. Vicidomini, M. Gulisano, A. Diaspro, L. Lanzanò, SPLIT-PIN software enabling confocal and super-resolution imaging with a virtually closed pinhole, Scientific reports 13, 2741 (2023).
L.-X. Gao, H. Hao, Y.-Q. Yu, J.-L. Chen, W.-Q. Chen, Z.-D. Gong, Y. Liu, F.-L. Jiang, Protein Labeling Facilitates the Understanding of Protein Corona Formation via Fluorescence Resonance Energy Transfer and Fluorescence Correlation Spectroscopy, Langmuir 39, 15275 (2023).
A. Gibney, R. de Paiva, V. Singh, R. Fox, D. Thompson, J. Hennessy, C. Slator, C. McKenzie, P. Johansson, V. McKee, F. Westerlund, A. Kellett, A Click Chemistry-Based Artificial Metallo-Nuclease, Angewandte Chemie International Edition 62, e202305759 (2023).
K. Hu, Q. Sun, R. Chen, T. Xu, Y. Li, L. Chen, A. Wang, H. Qi, D. Shao, H. Yue, Y. Wang, Z. Tang, Y. Wang, C. Liu, H. Lv, F. Wang, H. Xu, Expanding the toolset of fluorescent covalent staining of biological samples by labeling carboxylate and phosphate groups, Journal of Biophotonics 16, e202300027 (2023).
C. Jayachandran, A. Ghosh, M. Prabhune, J. Bath, A. Turberfield, L. Hauke, J. Enderlein, F. Rehfeldt, C. Schmidt, DNA-Based Optical Sensors for Forces in Cytoskeletal Networks, ACS Applied Nano Materials 6, 15455 (2023).
D. Kylies, M. Zimmermann, F. Haas, M. Schwerk, M. Kuehl, M. Brehler, J. Czogalla, L. Hernandez, L. Konczalla, Y. Okabayashi, J. Menzel, I. Edenhofer, S. Mezher, H. Aypek, B. Dumoulin, H. Wu, S. Hofmann, O. Kretz, N. Wanner, N. Tomas, S. Krasemann, M. Glatzel, C. Kuppe, R. Kramann, B. Banjanin, R. Schneider, C. Urbschat, P. Arck, N. Gagliani, M. van Zandvoort, T. Wiech, F. Grahammer, P. Sáez, M. Wong, S. Bonn, T. Huber, V. Puelles, Expansion-enhanced super-resolution radial fluctuations enable nanoscale molecular profiling of pathology specimens, Nature nanotechnology 18, 336 (2023).
Y. Li, Y. Niu, C. Kong, Z. Yang, J. Qu, Theoretical insight on the saturated stimulated emission intensity of a squaraine dye for STED nanoscopy, Spectrochimica Acta Part A: Molecular and Biomolecular Spectroscopy 284, 121793 (2023).
R. Lira, J. Hammond, R. Cavalcanti, M. Rous, K. Riske, W. Roos, The underlying mechanical properties of membranes tune their ability to fuse, Journal of Biological Chemistry 299, 105430 (2023).
A. Mangiarotti, N. Chen, Z. Zhao, R. Lipowsky, R. Dimova, Wetting and complex remodeling of membranes by biomolecular condensates, Nature communications 14, 2809 (2023).
D. Newman, L. Young, T. Waring, L. Brown, K. Wolanska, E. MacDonald, A. Charles-Orszag, B. Goult, P. Caswell, T. Sakuma, T. Yamamoto, L. Machesky, M. Morgan, T. Zech, 3D matrix adhesion feedback controls nuclear force coupling to drive invasive cell migration, Cell Reports 42, 113554 (2023).
N. Ponzar, N. Pozzi, Probing the conformational dynamics of thiol-isomerases using non-canonical amino acids and single-molecule FRET, Methods 214, 8 (2023).
R. Riera, E. Archontakis, G. Cremers, T. de Greef, P. Zijlstra, L. Albertazzi, Precision and Accuracy of Receptor Quantification on Synthetic and Biological Surfaces Using DNA-PAINT, ACS sensors 8, 80 (2023).
E. Salibi, B. Peter, P. Schwille, H. Mutschler, Periodic temperature changes drive the proliferation of self-replicating
R. Schmid, J. Kaiser, R. Willbold, N. Walther, R. Wittig, M. Lindén, Towards a simple in vitro surface chemistry pre-screening method for nanoparticles to be used for drug delivery to solid tumours, Biomaterials Science 11, 6287 (2023).
R. Schmid, M. Volcic, S. Fischer, Z. Qu, H. Barth, A. Popat, F. Kirchhoff, M. Lindén, Surface functionalization affects the retention and bio-distribution of orally administered mesoporous silica nanoparticles in a colitis mouse model, Scientific reports 13, 20175 (2023).
T. Sheard, T. Shakespeare, R. Seehra, M. Spencer, K. Suen, I. Jayasinghe, Differential labelling of human sub-cellular compartments with fluorescent dye esters and expansion microscopy, Nanoscale 15, 18489 (2023).
B. Sohmen, C. Beck, V. Frank, T. Seydel, I. Hoffmann, B. Hermann, M. Nüesch, M. Grimaldo, F. Schreiber, S. Wolf, F. Roosen-Runge, T. Hugel, The Onset of Molecule-Spanning Dynamics in Heat Shock Protein Hsp90, Environmental Toxicology 10, 2304262 (2023).
E. Soprano, M. Migliavacca, M. López-Ferreiro, B. Pelaz, E. Polo, P. del Pino, Fusogenic Cell-Derived nanocarriers for cytosolic delivery of cargo inside living cells, Journal of Colloid and Interface Science 648, 488 (2023).
F. Steiert, P. Schultz, S. Höfinger, T. Müller, P. Schwille, T. Weidemann, Insights into receptor structure and dynamics at the surface of living cells, Nature communications 14, 1596 (2023).
G. Wen, M. Lycas, Y. Jia, V. Leen, M. Sauer, J. Hofkens, Trifunctional Linkers Enable Improved Visualization of Actin by Expansion Microscopy, ACS Nano 17, 20589 (2023).
J. Wolff, L. Scheiderer, T. Engelhardt, J. Engelhardt, J. Matthias, S. Hell, MINFLUX dissects the unimpeded walking of kinesin-1, Science 379, 1004 (2023).
J. Xue, M. Zhang, J. Yong, Q. Chen, J. Wang, J. Xu, K. Liang, Light-Switchable Biocatalytic Covalent–Organic Framework Nanomotors for Aqueous Contaminants Removal, Nano Letters 23, 11243 (2023).
A. Yeşilyurt, M. Sanz-Paz, F. Zhu, X. Wu, K. Sunil, G. Acuna, J.-S. Huang, Unidirectional Meta-Emitters Based on the Kerker Condition Assembled by DNA Origami, ACS Nano 17, 19189 (2023).
J. Zähringer, F. Cole, J. Bohlen, F. Steiner, I. Kamińska, P. Tinnefeld, Combining pMINFLUX, graphene energy transfer and DNA-PAINT for nanometer precise 3D super-resolution microscopy, Light: Science & Applications 12, 1 (2023).
J. Zou, K. Mitra, P. Anees, D. Oettinger, J. Ramirez, A. Veetil, P. Gupta, R. Rao, J. Smith, P. Kratsios, Y. Krishnan, A DNA nanodevice for mapping sodium at single-organelle resolution, Nature biotechnology (2023).
M. Alsamsam, A. Kopūstas, M. Jurevičiūtė, M. Tutkus, The miEye, HardwareX 12, e00368 (2022).
S. An, M. Stagi, T. Gould, Y. Wu, M. Mlodzianoski, F. Rivera-Molina, D. Toomre, S. Strittmatter, P. de Camilli, J. Bewersdorf, D. Zenisek, Multimodal imaging of synaptic vesicles with a single probe, Cell Reports Methods 2, 100199 (2022).
M. Arista-Romero, P. Delcanale, S. Pujals, L. Albertazzi, Nanoscale Mapping of Recombinant Viral Proteins, ACS Photonics 9, 101 (2022).
J. Bachman, C. Wight, A. Bardo, A. Johnson, C. Pavlich, A. Boley, H. Wagner, J. Swaminathan, B. Iverson, E. Marcotte, E. Anslyn, Evaluating the Effect of Dye-Dye Interactions of Xanthene-Based Fluorophores in the Fluorosequencing of Peptides, Bioconjugate Chemistry 33, 1156 (2022).
Y. Bagheri, A. Ali, P. Keshri, J. Chambers, A. Gershenson, M. You, Imaging Membrane Order and Dynamic Interactions in Living Cells with a DNA Zipper Probe, Angewandte Chemie International Edition 61, e202112033 (2022).
A. Blanchard, Z. Li, E. Duran, C. Scull, J. Hoff, K. Wright, V. Pan, N. Walter, Ultra-photostable DNA FluoroCubes, Nano letters 22, 6235 (2022).
S. Chatterjee, R. Molenaar, W. de Vos, H. Roesink, R. Wagterveld, J. Cornelissen, Claessens, Mireille M. A. E., C. Blum, Quantification of the Retention and Disassembly of Virus Particles by a PEI-Functionalized Microfiltration Membrane, ACS applied polymer materials 4, 5173 (2022).
M. Gerstenberg, C. Stürzel, T. Weil, F. Kirchhoff, M. Lindén, Modular Hydrogel−Mesoporous Silica Nanoparticle Constructs for Therapy and Diagnostics, Advanced NanoBiomed Research 2, 2100125 (2022).
M. Gonzalez Pisfil, I. Nadelson, B. Bergner, S. Rottmeier, A. Thomae, S. Dietzel, Stimulated emission depletion microscopy with a single depletion laser using five fluorochromes and fluorescence lifetime phasor separation, Scientific Reports 12, 1 (2022).
A. Hancock, D. Swainsbury, S. Meredith, K. Morigaki, C. Hunter, P. Adams, Enhancing the spectral range of plant and bacterial light-harvesting pigment-protein complexes with various synthetic chromophores incorporated into lipid vesicles, Journal of Photochemistry and Photobiology B: Biology 237, 112585 (2022).
P. Harris, A. Narducci, C. Gebhardt, T. Cordes, S. Weiss, E. Lerner, Multi-parameter photon-by-photon hidden Markov modeling, Nature Communications 13, 1 (2022).
S. Hernández-Pérez, P. Mattila, A specific hybridisation internalisation probe (SHIP) enables precise live-cell and super-resolution imaging of internalized cargo, Scientific Reports 12, 620 (2022).
K. Hübner, M. Raab, J. Bohlen, J. Bauer, P. Tinnefeld, Salt-induced conformational switching of a flat rectangular DNA origami structure, Nanoscale 14, 7898 (2022).
R. Ibusuki, T. Morishita, A. Furuta, S. Nakayama, M. Yoshio, H. Kojima, K. Oiwa, K.’y. Furuta, Programmable molecular transport achieved by engineering protein motors to move on DNA nanotubes, Science 375, 1159 (2022).
H. Im, E. Heo, D.-H. Song, J. Park, H. Park, K. Kang, J.-B. Chang, Fabrication of heterogeneous chemical patterns on stretchable hydrogels using single-photon lithography, Soft Matter 18, 4402 (2022).
L. Jensen, S. Rao, M. Schuschnig, A. Cada, S. Martens, G. Hummer, J. Hurley, Membrane curvature sensing and stabilization by the autophagic LC3 lipidation machinery, Science Advances 8, eadd1436 (2022).
Y. Jin, J. Bae, T. Kim, H. Hwang, T. Kim, M. Yu, H. Oh, K. Hashiya, T. Bando, H. Sugiyama, K. Jo, Twelve Colors of Streptavidin-Fluorescent Proteins (SA-FPs), Analytical Chemistry 94, 16927 (2022).
P. Klier, R. Roo, E. Miller, Fluorescent indicators for imaging membrane potential of organelles, Current Opinion in Chemical Biology 71, 102203 (2022).
H. Mai, Z. Rong, S. Zhao, R. Cai, H. Steinke, I. Bechmann, A. Ertürk, Scalable tissue labeling and clearing of intact human organs, Nature Protocols 17, 2188 (2022).
A. Mota, M. Schweitzer, E. Wernersson, N. Crosetto, M. Bienko, Simultaneous visualization of DNA loci in single cells by combinatorial multi-color iFISH, Scientific Data 9, 1 (2022).
S. Mukherjee, J.-M. Knop, R. Winter, Modulation of the Conformational Space of SARS-CoV-2 RNA Quadruplex RG-1 by Cellular Components and the Amyloidogenic Peptides α-Synuclein and hIAPP, Chemistry – A European Journal 28, e202104182 (2022).
F. Muzzopappa, J. Hummert, M. Anfossi, S. Tashev, D.-P. Herten, F. Erdel, Detecting and quantifying liquid–liquid phase separation in living cells by model-free calibrated half-bleaching, Nature Communications 13, 1 (2022).
B. Nguyen, N. Tufenkji, Single-Particle Resolution Fluorescence Microscopy of Nanoplastics, Environmental Science & Technology (2022).
U. Pramanik, A. Nandy, L. Khamari, S. Mukherjee, Structure and Transition Dynamics of Intrinsically Disordered Proteins Probed by Single-Molecule Spectroscopy, Langmuir : the ACS journal of surfaces and colloids 38, 12764 (2022).
S. Puza, S. Caesar, C. Poojari, M. Jung, R. Seemann, J. Hub, B. Schrul, J.-B. Fleury, Lipid Droplets Embedded in a Model Cell Membrane Create a Phospholipid Diffusion Barrier, Small 18, 2106524 (2022).
E. Reville, E. Sylvester, S. Benware, S. Negi, E. Berda, Customizable molecular recognition, Polymer Chemistry 13, 3387 (2022).
R. Riera, E. Archontakis, G. Cremers, T. de Greef, P. Zijlstra, L. Albertazzi, Precision and Accuracy of Receptor Quantification on Synthetic and Biological Surfaces Using DNA-PAINT, ACS sensors 8, 80 (2023).
M. Safar, A. Saurabh, B. Sarkar, M. Fazel, K. Ishii, T. Tahara, I. Sgouralis, S. Pressé, Single-photon smFRET. III. Application to pulsed illumination, Biophysical Reports 2, 100088 (2022).
A. Saurabh, S. Niekamp, I. Sgouralis, S. Pressé, Modeling Non-additive Effects in Neighboring Chemically Identical Fluorophores, The journal of physical chemistry. B (2022).
E. Steib, R. Tetley, R. Laine, D. Norris, Y. Mao, J. Vermot, TissUExM enables quantitative ultrastructural analysis in whole vertebrate embryos by expansion microscopy, Cell Reports Methods 2, 100311 (2022).
K. Trofymchuk, K. Kołątaj, V. Glembockyte, F. Zhu, G. Acuna, T. Liedl, P. Tinnefeld, Gold Nanorod DNA Origami Antennas for 3 Orders of Magnitude Fluorescence Enhancement in NIR, ACS Nano (2023).
Y. Wang, J. Lin, Q. Zhang, X. Chen, H. Luan, M. Gu, Fluorescence Nanoscopy in Neuroscience, Engineering 16, 29 (2022).
M. Weber, H. von der Emde, M. Leutenegger, P. Gunkel, S. Sambandan, T. Khan, J. Keller-Findeisen, V. Cordes, S. Hell, MINSTED nanoscopy enters the Ångström localization range, Nature Biotechnology, 1 (2022).
J. Wolff, L. Scheiderer, T. Engelhardt, J. Engelhardt, J. Matthias, S. Hell, MINFLUX dissects the unimpeded walking of kinesin-1, Science 379, 1004 (2023).
C. Wu, T. Dougan, D. Walt, High-Throughput, High-Multiplex Digital Protein Detection with Attomolar Sensitivity, ACS Nano 16, 1025 (2022).
J. Zähringer, F. Cole, J. Bohlen, F. Steiner, I. Kamińska, P. Tinnefeld, Combining pMINFLUX, graphene energy transfer and DNA-PAINT for nanometer precise 3D super-resolution microscopy, Light: Science & Applications 12, 1 (2023).
J. Alston, A. Soranno, A. Holehouse, Integrating single-molecule spectroscopy and simulations for the study of intrinsically disordered proteins, Methods 193, 116 (2021).
X. An, S. Erramilli, B. Reinhard, Plasmonic nano-antimicrobials, Nanoscale 13, 3374 (2021).
T. Andrian, P. Delcanale, S. Pujals, L. Albertazzi, Correlating Super-Resolution Microscopy and Transmission Electron Microscopy Reveals Multiparametric Heterogeneity in Nanoparticles, Nano Letters 21, 5360 (2021).
T. Andrian, S. Pujals, L. Albertazzi, Quantifying the effect of PEG architecture on nanoparticle ligand availability using DNA-PAINT, Nanoscale Advances 3, 6876 (2021).
D. Bhatia, C. Wunder, L. Johannes, Self-assembled, Programmable DNA Nanodevices for Biological and Biomedical Applications, ChemBioChem 22, 763 (2021).
B. Cao, S. Coelho, J. Li, G. Wang, A. Pertsinidis, Volumetric interferometric lattice light-sheet imaging, Nature Biotechnology 39, 1385 (2021).
X. Chen, T. Liu, X. Qin, Q. Nguyen, S. Lee, C. Lee, Y. Ren, J. Chu, G. Zhu, T.-Y. Yoon, C. Park, H. Park, Simultaneous Real-Time Three-Dimensional Localization and FRET Measurement of Two Distinct Particles, Nano Letters 21, 7479 (2021).
C. Cheubong, E. Takano, Y. Kitayama, H. Sunayama, K. Minamoto, R. Takeuchi, S. Furutani, T. Takeuchi, Molecularly imprinted polymer nanogel-based fluorescence sensing of pork contamination in halal meat extracts, Biosensors and Bioelectronics 172, 112775 (2021).
M. Chinnaraj, D. Barrios, C. Frieden, T. Heyduk, R. Flaumenhaft, N. Pozzi, Bioorthogonal Chemistry Enables Single-Molecule FRET Measurements of Catalytically Active Protein Disulfide Isomerase, Encyclopedia of Analytical Chemistry 22, 134 (2021).
L. Cruz-Zaragoza, S. Dennerlein, A. Linden, R. Yousefi, E. Lavdovskaia, A. Aich, R. Falk, R. Gomkale, T. Schöndorf, M. Bohnsack, R. Richter-Dennerlein, H. Urlaub, P. Rehling, An in vitro system to silence mitochondrial gene expression, Cell 184, 5824-5837.e15 (2021).
L. Doll, J. Lackner, F. Rönicke, G. Nienhaus, H.-A. Wagenknecht, Fluorescence Lifetime Imaging Microscopy (FLIM) of Intracellular Transport by Means of Doubly Labelled siRNA Architectures, ChemBioChem 22, 2561 (2021).
M. Ganji, T. Schlichthaerle, A. Eklund, S. Strauss, R. Jungmann, Quantitative assessment of labeling probes for super‐resolution microscopy using designer DNA nanostructures, ChemPhysChem (2021).
M. Gonzalez Pisfil, S. Rohilla, M. König, B. Krämer, M. Patting, F. Koberling, R. Erdmann, Triple-Color STED Nanoscopy, The Journal of Physical Chemistry B 125, 5694 (2021).
C. Götz, G. Hinze, A. Gellert, H. Maus, F. von Hammerstein, S. Hammerschmidt, L. Lauth, U. Hellmich, T. Schirmeister, T. Basché, Conformational Dynamics of the Dengue Virus Protease Revealed by Fluorescence Correlation and Single-Molecule FRET Studies, The Journal of Physical Chemistry B 125, 6837 (2021).
C. Hinson, A. Bardo, C. Shannon, S. Rivera, J. Swaminathan, E. Marcotte, E. Anslyn, Studies of Surface Preparation for the Fluorosequencing of Peptides, Langmuir 37, 14856 (2021).
S. Horvat, Y. Yu, H. Manz, T. Keller, A. Beilhack, J. Groll, K. Albrecht, Nanogels as Antifungal-Drug Delivery System Against Aspergillus Fumigatus, Advanced NanoBiomed Research 1, 2000060 (2021).
G. Kasagi, Y. Yoneda, M. Kondo, H. Miyasaka, Y. Nagasawa, T. Dewa, Enhanced light harvesting and photocurrent generation activities of biohybrid light–harvesting 1–reaction center core complexes (LH1-RCs) from Rhodopseudomonas palustris, Journal of Photochemistry and Photobiology A: Chemistry 405, 112790 (2021).
W. Liu, Q. Qiao, J. Zheng, J. Chen, W. Zhou, N. Xu, J. Li, L. Miao, Z. Xu, An assembly-regulated SNAP-tag fluorogenic probe for long-term super-resolution imaging of mitochondrial dynamics, Biosensors and Bioelectronics 176, 112886 (2021).
Y. Long, K. Ubych, E. Jagu, R. Neely, FRET-Based Method for Direct, Real-Time Measurement of DNA Methyltransferase Activity, Bioconjugate Chemistry 32, 192 (2021).
M. Mitsui, Y. Takakura, K. Hirata, Y. Niihori, Y. Fujiwara, K. Kobayashi, Excited-State Symmetry Breaking in a Multiple Multipolar Chromophore Probed by Single-Molecule Fluorescence Imaging and Spectroscopy, The Journal of Physical Chemistry B 125, 9950 (2021).
M. Pfeiffer, K. Trofymchuk, S. Ranallo, F. Ricci, F. Steiner, F. Cole, V. Glembockyte, P. Tinnefeld, Single antibody detection in a DNA origami nanoantenna, iScience 24, 103072 (2021).
A. Priyadarshi, F. Dullo, D. Wolfson, A. Ahmad, N. Jayakumar, V. Dubey, J.-C. Tinguely, B. Ahluwalia, G. Murugan, A transparent waveguide chip for versatile total internal reflection fluorescence-based microscopy and nanoscopy, Communications Materials 2, 1 (2021).
T. Schröder, S. Bange, J. Schedlbauer, F. Steiner, J. Lupton, P. Tinnefeld, J. Vogelsang, How Blinking Affects Photon Correlations in Multichromophoric Nanoparticles, ACS Nano 15, 18037 (2021).
M. Sunbul, J. Lackner, A. Martin, D. Englert, B. Hacene, F. Grün, K. Nienhaus, G. Nienhaus, A. Jäschke, Super-resolution RNA imaging using a rhodamine-binding aptamer with fast exchange kinetics, Nature Biotechnology 39, 686 (2021).
A. Szalai, C. Zaza, F. Stefani, Super-resolution FRET measurements, Nanoscale 13, 18421 (2021).
S. Thill, T. Schmidt, D. Wöll, R. Gebhardt, A regenerated fiber from rennet-treated casein micelles, Colloid and Polymer Science 15, 201 (2021).
K. Tsutsumi, H. Sunayama, Y. Kitayama, E. Takano, Y. Nakamachi, R. Sasaki, T. Takeuchi, Fluorescent Signaling of Molecularly Imprinted Nanogels Prepared via Postimprinting Modifications for Specific Protein Detection, Advanced NanoBiomed Research 1, 2000079 (2021).
M. Weber, M. Leutenegger, S. Stoldt, S. Jakobs, T. Mihaila, A. Butkevich, S. Hell, MINSTED fluorescence localization and nanoscopy, Nature Photonics 19, 780 (2021).
H. Wei, X. Yan, Y. Niu, Q. Li, Z. Jia, H. Xu, Plasmon–Exciton Interactions, Advanced Functional Materials 31, 2100889 (2021).
J. Weiden, M. Schluck, M. Ioannidis, van Dinther, Eric A. W., M. Rezaeeyazdi, F. Omar, J. Steuten, D. Voerman, J. Tel, M. Diken, S. Bencherif, C. Figdor, M. Verdoes, Robust Antigen-Specific T Cell Activation within Injectable 3D Synthetic Nanovaccine Depots, ACS Biomaterials Science & Engineering 7, 5622 (2021).
Y. Wu, X. Han, Y. Su, M. Glidewell, J. Daniels, J. Liu, T. Sengupta, I. Rey-Suarez, R. Fischer, A. Patel, C. Combs, J. Sun, X. Wu, R. Christensen, C. Smith, L. Bao, Y. Sun, L. Duncan, J. Chen, Y. Pommier, Y.-B. Shi, E. Murphy, S. Roy, A. Upadhyaya, D. Colón-Ramos, P. La Riviere, H. Shroff, Multiview confocal super-resolution microscopy, Nature 600, 279 (2021).
A. Zengin, J. Castro, P. Habibovic, S. van Rijt, Injectable, self-healing mesoporous silica nanocomposite hydrogels with improved mechanical properties, Nanoscale 13, 1144 (2021).
V. Zila, E. Margiotta, B. Turoňová, T. Müller, C. Zimmerli, S. Mattei, M. Allegretti, K. Börner, J. Rada, B. Müller, M. Lusic, H.-G. Kräusslich, M. Beck, Cone-shaped HIV-1 capsids are transported through intact nuclear pores, Cell (2021).
A. Al Masud, W. Martin, F. Moonschi, S. Park, B. Srijanto, K. Graham, C. Collier, C. Richards, Mixed metal zero-mode guides (ZMWs) for tunable fluorescence enhancement, Nanoscale Advances 2, 1894 (2020).
P. Bacchin, D. Snisarenko, D. Stamatialis, P. Aimar, C. Causserand, Combining fluorescence and permeability measurements in a membrane microfluidic device to study protein sorption mechanisms, Journal of Membrane Science 614, 118485 (2020).
W.-C. Chen, Y. Simanjuntak, L.-W. Chu, Y.-H. Ping, Y.-L. Lee, Y.-L. Lin, W.-S. Li, Benzenesulfonamide Derivatives as Calcium/Calmodulin-Dependent Protein Kinase Inhibitors and Antiviral Agents against Dengue and Zika Virus Infections, Journal of Medicinal Chemistry 63, 1313 (2020).
Eugene Kim, Jacob Kerssemakers, Indra A. Shaltiel, Christian H. Haering, Cees Dekker, DNA-loop extruding condensin complexes can traverse one another, Nature 579, 438 (2020).
R. Götz, T. Kunz, J. Fink, F. Solger, J. Schlegel, J. Seibel, V. Kozjak-Pavlovic, T. Rudel, M. Sauer, Nanoscale imaging of bacterial infections by sphingolipid expansion microscopy, Nature Communications 11, 7478 (2020).
R. Ibusuki, M. Shiraga, A. Furuta, M. Yoshio, H. Kojima, K. Oiwa, K.’y. Furuta, Collective motility of dynein linear arrays built on DNA nanotubes, Biochemical and Biophysical Research Communications 523, 1014 (2020).
M. Jäpel, F. Gerth, T. Sakaba, J. Bacetic, L. Yao, S.-J. Koo, T. Maritzen, C. Freund, V. Haucke, Intersectin-Mediated Clearance of SNARE Complexes Is Required for Fast Neurotransmission, Cell Reports 30, 409-420.e6 (2020).
D. Jullié, M. Stoeber, J.-B. Sibarita, H. Zieger, T. Bartol, S. Arttamangkul, T. Sejnowski, E. Hosy, M. von Zastrow, A Discrete Presynaptic Vesicle Cycle for Neuromodulator Receptors, Neuron 105, 663-677.e8 (2020).
S. Koch, A.-B. Seinen, M. Kamel, D. Kuckla, C. Monzel, A. Kedrov, A. Driessen, Single-molecule analysis of dynamics and interactions of the SecYEG translocon, The FEBS Journal 228, 2203 (2020).
T. Köhler, T. Heida, S. Hoefgen, N. Weigel, V. Valiante, J. Thiele, Cell-free protein synthesis and in situ immobilization of deGFP-MatB in polymer microgels for malonate-to-malonyl CoA conversion, RSC Advances 10, 40588 (2020).
K. Kramm, T. Schröder, J. Gouge, A. Vera, K. Gupta, F. Heiss, T. Liedl, C. Engel, I. Berger, A. Vannini, P. Tinnefeld, D. Grohmann, DNA origami-based single-molecule force spectroscopy elucidates RNA Polymerase III pre-initiation complex stability, Nature Communications 11, 2828 (2020).
X. Li, J. Zheng, W. Liu, Q. Qiao, J. Chen, W. Zhou, Z. Xu, Long-term super-resolution imaging of mitochondrial dynamics, Chinese Chemical Letters 31, 2937 (2020).
M. Liebel, J. Ortega Arroyo, V. Beltrán, J. Osmond, A. Jo, H. Lee, R. Quidant, N. van Hulst, 3D tracking of extracellular vesicles by holographic fluorescence imaging, Science Advances 6 (2020).
J. Liu, Z. Guo, M. Kordanovski, J. Kaltbeitzel, H. Zhang, Z. Cao, Z. Gu, P. Wich, M. Lord, K. Liang, Metal-organic frameworks as protective matrices for peptide therapeutics, Journal of Colloid and Interface Science 576, 356 (2020).
J. Liu, T. Hebbrecht, T. Brans, E. Parthoens, S. Lippens, C. Li, H. de Keersmaecker, W. de Vos, S. de Smedt, R. Boukherroub, J. Gettemans, R. Xiong, K. Braeckmans, Long-term live-cell microscopy with labeled nanobodies delivered by laser-induced photoporation, Nano Research 13, 485 (2020).
D. Mai, C. Schroeder, 100th Anniversary of Macromolecular Science Viewpoint, ACS Macro Letters 9, 1332 (2020).
C. Mao, M. Lee, J.-R. Jhan, A. Halpern, M. Woodworth, A. Glaser, T. Chozinski, L. Shin, J. Pippin, S. Shankland, J. Liu, J. Vaughan, Feature-rich covalent stains for super-resolution and cleared tissue fluorescence microscopy, Science Advances 6, eaba4542 (2020).
H. Mazloom-Farsibaf, W. Kanagy, D. Lidke, K. Lidke, High-speed single molecule imaging datasets of membrane proteins in rat basophilic leukemia cells, Data in brief 30, 105424 (2020).
S. Mukherjee, J.-M. Knop, S. Möbitz, R. Winter, Alteration of the Conformational Dynamics of a DNA Hairpin by α-Synuclein in the Presence of Aqueous Two-Phase Systems, Chemistry – A European Journal 26, 10987 (2020).
S. Nummelin, B. Shen, P. Piskunen, Q. Liu, M. Kostiainen, V. Linko, Robotic DNA Nanostructures, ACS synthetic biology (2020).
B. Plochberger, T. Sych, F. Weber, J. Novacek, M. Axmann, H. Stangl, E. Sezgin, Lipoprotein Particles Interact with Membranes and Transfer Their Cargo without Receptors, Biochemistry 59, 4421 (2020).
A. Rae, X. Wei, N. Flores-Rodriguez, D. McCurdy, D. Collings, Super-Resolution Fluorescence Imaging of Arabidopsis thaliana Transfer Cell Wall Ingrowths using Pseudo-Schiff Labelling Adapted for the Use of Different Dyes, Plant & cell physiology 61, 1775 (2020).
A. Rajwar, V. Morya, S. Kharbanda, D. Bhatia, DNA Nanodevices to Probe and Program Membrane Organization, Dynamics, and Applications, The Journal of membrane biology 253, 577 (2020).
S. Reshetniak, J.-E. Ußling, E. Perego, B. Rammner, T. Schikorski, E. Fornasiero, S. Truckenbrodt, S. Köster, S. Rizzoli, A comparative analysis of the mobility of 45 proteins in the synaptic bouton, The EMBO Journal 39, e104596 (2020).
Ł. Richter, P. Albrycht, M. Księżopolska-Gocalska, E. Poboży, R. Bachliński, V. Sashuk, J. Paczesny, R. Hołyst, Fast and efficient deposition of broad range of analytes on substrates for surface enhanced Raman spectroscopy, Biosensors and Bioelectronics 156, 112124 (2020).
C. Robidillo, J. Veinot, Functional Bio-inorganic Hybrids from Silicon Quantum Dots and Biological Molecules, ACS Applied Materials & Interfaces 12, 52251 (2020).
P. Sharma, S. Parthasarathi, N. Patil, M. Waskar, J. Raut, M. Puranik, K. Ayappa, J. Basu, Assessing Barriers for Antimicrobial Penetration in Complex Asymmetric Bacterial Membranes, Langmuir (2020).
B. Shen, P. Piskunen, S. Nummelin, Q. Liu, M. Kostiainen, V. Linko, Advanced DNA Nanopore Technologies, ACS Applied Bio Materials 3, 5606 (2020).
Y. Shinsato, A. Doyle, W. Li, K. Yamada, Direct comparison of five different 3D extracellular matrix model systems for characterization of cancer cell migration, Cancer Reports 3, e1257 (2020).
M. Singh, M. Watkinson, E. Scanlan, G. Miller, Illuminating glycoscience, RSC Chemical Biology 1, 352 (2020).
V. Singh, P. Johansson, D. Torchinsky, Y.-L. Lin, R. Öz, Y. Ebenstein, O. Hammarsten, F. Westerlund, Quantifying DNA damage induced by ionizing radiation and hyperthermia using single DNA molecule imaging, Translational Oncology 13, 100822 (2020).
Sophie Massou, Filipe Nunes Vicente, Franziska Wetzel, Amine Mehidi, Dan Strehle, Cecile Leduc, Raphaël Voituriez, Olivier Rossier, Pierre Nassoy, Grégory Giannone, Cell stretching is amplified by active actin remodelling to deform and recruit proteins in mechanosensitive structures, Nature Cell Biology 22, 1011 (2020).
B. Stinson, A. Moreno, J. Walter, J. Loparo, A Mechanism to Minimize Errors during Non-homologous End Joining, Molecular Cell 77, 1080-1091.e8 (2020).
S. Thill, T. Schmidt, D. Wöll, R. Gebhardt, Single particle tracking as a new tool to characterise the rennet coagulation process, International Dairy Journal 105, 104659 (2020).
S. Varela-Aramburu, C. Ghosh, F. Goerdeler, P. Priegue, O. Moscovitz, P. Seeberger, Targeting and Inhibiting Plasmodium falciparum Using Ultra-small Gold Nanoparticles, ACS Applied Materials & Interfaces 12, 43380 (2020).
L. Villegas-Hernández, M. Nystad, F. Ströhl, P. Basnet, G. Acharya, B. Ahluwalia, Visualizing ultrastructural details of placental tissue with super-resolution structured illumination microscopy, Placenta 97, 42 (2020).
Y. Wang, M. Horáček, P. Zijlstra, Strong Plasmon Enhancement of the Saturation Photon Count Rate of Single Molecules, The journal of physical chemistry letters 11, 1962 (2020).
C. Wu, P. Garden, D. Walt, Ultrasensitive Detection of Attomolar Protein Concentrations by Dropcast Single Molecule Assays, Journal of the American Chemical Society 142, 12314 (2020).
K. Bielec, G. Bubak, T. Kalwarczyk, R. Holyst, Analysis of Brightness of a Single Fluorophore for Quantitative Characterization of Biochemical Reactions, The Journal of Physical Chemistry B 124, 1941 (2020).
M. Gast, F. Wondany, B. Raabe, J. Michaelis, H. Sobek, B. Mizaikoff, Use of Super-Resolution Optical Microscopy To Reveal Direct Virus Binding at Hybrid Core-Shell Matrixes, Analytical Chemistry 92, 3050 (2020).
J. Kuhn, Y. Lin, A. Krhac Levacic, N. Al Danaf, L. Peng, M. Höhn, D. Lamb, E. Wagner, U. Lächelt, Delivery of Cas9/sgRNA Ribonucleoprotein Complexes via Hydroxystearyl Oligoamino Amides, Bioconjugate Chemistry 31, 729 (2020).
N. Melnychuk, S. Egloff, A. Runser, A. Reisch, A. Klymchenko, Light‐Harvesting Nanoparticle Probes for FRET‐Based Detection of Oligonucleotides with Single‐Molecule Sensitivity, Angewandte Chemie International Edition 59, 6811 (2020).
L. Metzler, U. Rehbein, J.-N. Schönberg, T. Brandstetter, K. Thedieck, J. Rühe, Breaking the Interface, Analytical Chemistry (2020).
U. Rehman, S. Das, Y.-F. Chen, F.-J. Kao, High temporal resolution and polarization resolved fluorescence lifetime measurements through stimulated emission, Methods and Applications in Fluorescence 8, 24008 (2020).
D. Roy, J. Steinkühler, Z. Zhao, R. Lipowsky, R. Dimova, Mechanical Tension of Biomembranes Can Be Measured by Super Resolution (STED) Microscopy of Force-Induced Nanotubes, Nano letters 20, 3185 (2020).
T. Sansen, D. Sanchez-Fuentes, R. Rathar, A. Colom-Diego, F. El Alaoui, J. Viaud, M. Macchione, S. de Rossi, S. Matile, R. Gaudin, V. Bäcker, A. Carretero-Genevrier, L. Picas, Mapping Cell Membrane Organization and Dynamics Using Soft Nanoimprint Lithography, ACS Applied Materials & Interfaces (2020).
A. Wilkinson, E. Jagu, K. Ubych, S. Coulthard, A. Rushton, J. Kennefick, Q. Su, R. Neely, P. Fernandez-Trillo, Site-Selective and Rewritable Labeling of DNA through Enzymatic, Reversible, and Click Chemistries, ACS Central Science 6, 525 (2020).
C. Xu, X.-Y. He, Y. Peng, B.-S. Dai, B.-Y. Liu, S.-X. Cheng, Facile Strategy To Enhance Specificity and Sensitivity of Molecular Beacons by an Aptamer-Functionalized Delivery Vector, Analytical Chemistry 92, 2088 (2020).
Y. Yoneda, A. Goto, N. Takeda, H. Harada, M. Kondo, H. Miyasaka, Y. Nagasawa, T. Dewa, Ultrafast Photodynamics and Quantitative Evaluation of Biohybrid Photosynthetic Antenna and Reaction Center Complexes Generating Photocurrent, The Journal of Physical Chemistry C 124, 8605 (2020).
A. Ahmadi, K. Till, Y. Hafting, M. Schüttpelz, M. Bjørås, K. Glette, J. Tørresen, A. Rowe, B. Dalhus, Additive manufacturing of laminar flow cells for single-molecule experiments, Scientific Reports 9 (2019).
M. Baibakov, S. Patra, J.-B. Claude, A. Moreau, J. Lumeau, J. Wenger, Extending Single-Molecule Förster Resonance Energy Transfer (FRET) Range beyond 10 Nanometers in Zero-Mode Waveguides, ACS Nano 13, 8469 (2019).
A. Barth, L. Voith von Voithenberg, D. Lamb, Quantitative Single-Molecule Three-Color Förster Resonance Energy Transfer by Photon Distribution Analysis, The journal of physical chemistry. B 123, 6901 (2019).
R. Batchelor, T. Messer, M. Hippler, M. Wegener, C. Barner-Kowollik, E. Blasco, Two in One, Advanced Materials 31, 1904085 (2019).
E. Björk, B. Baumann, F. Hausladen, R. Wittig, M. Lindén, Cell adherence and drug delivery from particle based mesoporous silica films, RSC Advances 9, 17745 (2019).
M. de Boer, G. Gouridis, R. Vietrov, S. Begg, G. Schuurman-Wolters, F. Husada, N. Eleftheriadis, B. Poolman, C. McDevitt, T. Cordes, Conformational and dynamic plasticity in substrate-binding proteins underlies selective transport in ABC importers 8, e44652 (2019).
Y. Choi, C. Schmidt, P. Tinnefeld, I. Bald, S. Rödiger, A new reporter design based on DNA origami nanostructures for quantification of short oligonucleotides using microbeads, Scientific Reports 9, 1 (2019).
S. Das, I.-C. Chen, K. Rehman, J.-L. Hsu, G.-Y. Zhuo, F.-J. Kao, Background free imaging in stimulated emission fluorescence microscopy, Journal of Optics 21, 125301 (2019).
S. Das, Y.-C. Liang, S. Tanaka, Y. Ozeki, F.-J. Kao, Synchronized subharmonic modulation in stimulated emission microscopy, Optics Express 27, 27159 (2019).
A. Ghosh, A. Sharma, A. Chizhik, S. Isbaner, D. Ruhlandt, R. Tsukanov, I. Gregor, N. Karedla, J. Enderlein, Graphene-based metal-induced energy transfer for sub-nanometre optical localization, Nature Photonics 313, 1642 (2019).
M. Goetzfried, K. Vogele, A. Mückl, M. Kaiser, N. Holland, F. Simmel, T. Pirzer, Periodic Operation of a Dynamic DNA Origami Structure Utilizing the Hydrophilic–Hydrophobic Phase‐Transition of Stimulus‐Sensitive Polypeptides, Small 15 (2019).
W. Gong, P. Das, S. Samanta, J. Xiong, W. Pan, Z. Gu, J. Zhang, J. Qu, Z. Yang, Redefining the photo-stability of common fluorophores with triplet state quenchers, Chemical Communications 55, 8695 (2019).
H. Höfig, O. Yukhnovets, C. Remes, N. Kempf, A. Katranidis, D. Kempe, J. Fitter, Brightness-gated two-color coincidence detection unravels two distinct mechanisms in bacterial protein translation initiation, Communications Biology 2, 1 (2019).
I. Iachina, I. Antonescu, J. Dreier, J. Sørensen, J. Brewer, The nanoscopic molecular pathway through human skin, Biochimica et Biophysica Acta (BBA) - General Subjects 1863, 1226 (2019).
S. Kempter, A. Khmelinskaia, M. Strauss, P. Schwille, R. Jungmann, T. Liedl, W. Bae, Single Particle Tracking and Super-Resolution Imaging of Membrane-Assisted Stop-and-Go Diffusion and Lattice Assembly of DNA Origami, ACS Nano 13, 996 (2019).
V.V.G. Krishna Inavalli, M. Lenz, C. Butler, J. Angibaud, B. Compans, F. Levet, J. Tønnesen, O. Rossier, G. Giannone, O. Thoumine, E. Hosy, D. Choquet, J.-B. Sibarita, U. Nägerl, A super-resolution platform for correlative live single-molecule imaging and STED microscopy, Nature Methods 16, 1263 (2019).
C. Léger, T. Di Meo, M. Aumont-Nicaise, C. Velours, D. Durand, Li de la Sierra-Gallay, Ines, H. van Tilbeurgh, N. Hildebrandt, M. Desmadril, A. Urvoas, M. Valerio-Lepiniec, P. Minard, Ligand-induced conformational switch in an artificial bidomain protein scaffold, Scientific Reports 9 (2019).
W. Li, Z. Tong, K. Xiao, Z. Liu, Q. Gao, J. Sun, S. Liu, S. Han, Z. Wang, Single-frame wide-field nanoscopy based on ghost imaging via sparsity constraints, Optica 6, 1515 (2019).
C.-Y. Lin, C.-M. Yang, M. Lindén, Influence of serum concentration and surface functionalization on the protein adsorption to mesoporous silica nanoparticles, RSC Advances 9, 33912 (2019).
J. Linarès-Loyez, J. Ferreira, O. Rossier, B. Lounis, G. Giannone, L. Groc, L. Cognet, P. Bon, Self-Interference (SELFI) Microscopy for Live Super-Resolution Imaging and Single Particle Tracking in 3D, Frontiers in Physics 7, 68 (2019).
Q. Liu, Y. Chen, W. Liu, Y. Han, R. Cao, Z. Zhang, C. Kuang, X. Liu, Total internal reflection fluorescence pattern-illuminated Fourier ptychographic microscopy, Optics and Lasers in Engineering 123, 45 (2019).
F. Mayer, S. Richter, J. Westhauser, E. Blasco, C. Barner-Kowollik, M. Wegener, Multimaterial 3D laser microprinting using an integrated microfluidic system, Science Advances 5, eaau9160 (2019).
M. de Ruiter, R. van der Hee, A.J.M. Driessen, E. Keurhorst, M. Hamid, J.J.L.M. Cornelissen, Polymorphic assembly of virus-capsid proteins around DNA and the cellular uptake of the resulting particles, Journal of Controlled Release 307, 342 (2019).
S. Samanta, Y. He, A. Sharma, J. Kim, W. Pan, Z. Yang, J. Li, W. Yan, L. Liu, J. Qu, J. Kim, Fluorescent Probes for Nanoscopic Imaging of Mitochondria, Chem 5, 1697 (2019).
T. Schröder, M. Scheible, F. Steiner, J. Vogelsang, P. Tinnefeld, Interchromophoric Interactions Determine the Maximum Brightness Density in DNA Origami Structures, Nano letters 19, 1275 (2019).
F. Schueder, J. Stein, F. Stehr, A. Auer, B. Sperl, M. Strauss, P. Schwille, R. Jungmann, An order of magnitude faster DNA-PAINT imaging by optimized sequence design and buffer conditions, Nature Methods 16, 1101 (2019).
M. Segal, A. Ingargiola, E. Lerner, S. Chung, J. White, A. Streets, S. Weiss, X. Michalet, High-throughput smFRET analysis of freely diffusing nucleic acid molecules and associated proteins, Methods 169, 21 (2019).
A.R. Sekhar, B. Mallik, V. Kumar, J. Sankar, A cell-permeant small molecule for the super-resolution imaging of the endoplasmic reticulum in live cells, Organic & Biomolecular Chemistry 17, 3732 (2019).
S. Shah, A. Dubey, J. Reif, Programming Temporal DNA Barcodes for Single-Molecule Fingerprinting, Nano letters 19, 2668 (2019).
M. Sivasubramanian, Y. Chuang, L.-W. Lo, Evolution of Nanoparticle-Mediated Photodynamic Therapy, Molecules 24, 520 (2019).
M. Stobiecka, K. Ratajczak, S. Jakiela, Toward early cancer detection, Biosensors and Bioelectronics 137, 58 (2019).
T. Vavrdová, O. Šamajová, P. Křenek, M. Ovečka, P. Floková, R. Šnaurová, J. Šamaj, G. Komis, Multicolour three dimensional structured illumination microscopy of immunolabeled plant microtubules and associated proteins, Plant Methods 15, 1 (2019).
H. Vijayamohanan, P. Bhide, D. Boyd, Z. Zhou, E. Palermo, C. Ullal, Effect of Chemical Microenvironment in Spirothiopyran Monolayer Direct-Write Photoresists, Langmuir 35, 3871 (2019).
H. Vijayamohanan, G. Kenath, E. Palermo, C. Ullal, Super-resolution interference lithography enabled by non-equilibrium kinetics of photochromic monolayers, RSC Advances 9, 28841 (2019).
C. Wu, P. Garden, D. Walt, Ultrasensitive Detection of Attomolar Protein Concentrations by Dropcast Single Molecule Assays, Journal of the American Chemical Society 142, 12314 (2020).
L. Xin, M. Lu, S. Both, M. Pfeiffer, M. Urban, C. Zhou, H. Yan, T. Weiss, N. Liu, K. Lindfors, Watching a Single Fluorophore Molecule Walk into a Plasmonic Hotspot, ACS Photonics 6, 985 (2019).
A. Zrehen, D. Huttner, A. Meller, On-Chip Stretching, Sorting, and Electro-Optical Nanopore Sensing of Ultralong Human Genomic DNA, ACS Nano 13, 14388 (2019).
R. Jurado, J. Adamcik, M. López-Haro, J. González-Vera, Á. Ruiz-Arias, A. Sánchez-Ferrer, R. Cuesta, J. Domínguez-Vera, J. Calvino, A. Orte, R. Mezzenga, N. Gálvez, Apoferritin Protein Amyloid Fibrils with Tunable Chirality and Polymorphism, Journal of the American Chemical Society 141, 1606 (2019).