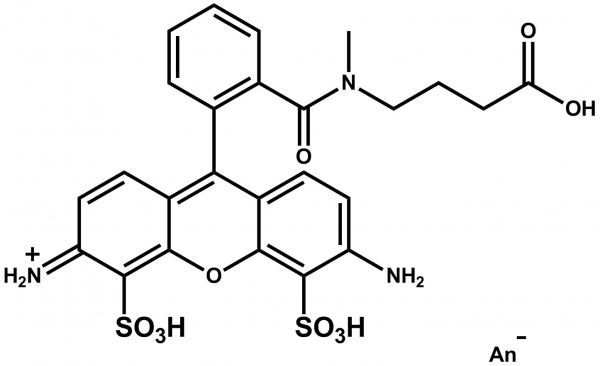
ATTO 488



|
Optische Eigenschaften |
||||
|
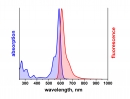
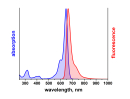
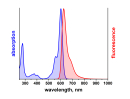
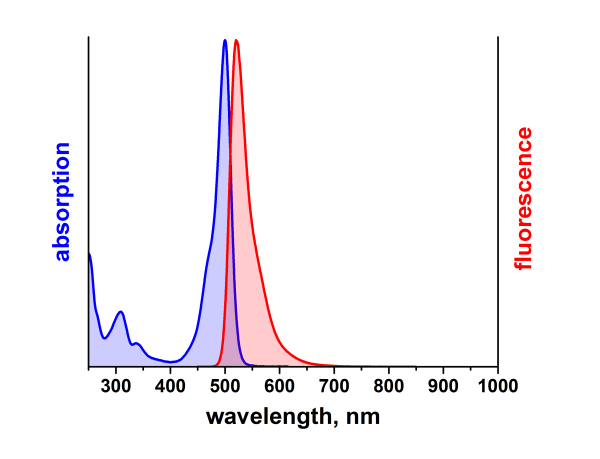
λabs |
500 |
nm |
||
|
εmax |
9,0×104 |
M-1 cm-1 |
||
|
λfl |
520 |
nm |
||
|
ηfl |
80 |
% |
||
|
τfl |
4,1 |
ns |
||
|
CF260 = ε260/εmax |
0,22 |
|
||
|
CF280 = ε280/εmax |
0,09 |
|
||
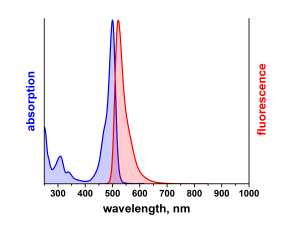
ATTO 488 ist ein hydrophiler Fluoreszenzmarker mit hervorragender Wasserlöslichkeit. Zu den charakteristischen Eigenschaften des Farbstoffs zählen starke Absorption, sehr hohe Fluoreszenzquantenausbeute sowie hohe thermische und photochemische Stabilität. Der Farbstoff ist somit besonders geeignet im Bereich der Einzelmoleküldetektion sowie zur hochauflösenden Mikroskopie (PALM, dSTORM, STED, etc.). Eine effiziente Anregung der Fluoreszenz erfolgt im Bereich 480 - 515 nm. Eine für den Farbstoff geeignete Anregungslichtquelle ist der Argon-Ionen-Laser mit seiner Linie bei 488 nm.
Produktdatenblätter
- ATTO 488 Produktinformation
- NHS-Ester
- Maleimid
- Iodacetamid
- Phalloidin
- Amin
- Azid/Alkin
- Streptavidin
Absorptions- und Emissionsspektrum (ASCII)
- ATTO 488 Absorption (.txt)
- ATTO 488 Emission (.txt)
Absorptions- und Emissionsspektrum (Grafik)
- ATTO 488 Absorption/Fluoreszenz (.jpg)
Sicherheitsdatenblätter
- ATTO 488 Carboxy MSDS
- ATTO 488 NHS-Ester MSDS
- ATTO 488 Maleimid MSDS
- ATTO 488 Streptavidin MSDS
- ATTO 488 Biotin MSDS
- ATTO 488 Phalloidin MSDS
- ATTO 488 Amin MSDS
- ATTO 488 Azid MSDS
- ATTO 488 Iodacetamid MSDS
- ATTO 488 Hydrazid MSDS
- ATTO 488 Alkin MSDS
- ATTO 488 Cadaverin MSDS
- ATTO 488 Tetrazin (MeTet) MSDS
- ATTO 488 Peg(4)-DBCO MSDS
Ausgewählte Literatur zu ATTO 488:
H. Alizadeh Zeinabad, W. Yeoh, M. Arif, M. Lomora, Y. Banz, C. Riether, P. Krebs, E. Szegezdi, Natural killer cell-mimic nanoparticles can actively target and kill acute myeloid leukemia cells, Biomaterials 298, 122126 (2023).
C. Alter, P. Detampel, R. Schefer, C. Lotter, P. Hauswirth, R. Puligilla, V. Weibel, S. Schenk, W. Heusermann, M. Schürz, N. Meisner-Kober, C. Palivan, T. Einfalt, J. Huwyler, High efficiency preparation of monodisperse plasma membrane derived extracellular vesicles for therapeutic applications, Communications biology 6, 478 (2023).
K. Augustyniak, A. Pragnaca, M. Lesniak, M. Halasa, A. Borkowska, E. Pieta, W. Kwiatek, C. Kieda, R. Zdanowski, K. Malek, Molecular tracking of interactions between progenitor and endothelial cells via Raman and FTIR spectroscopy imaging, Cellular and molecular life sciences : CMLS 80, 329 (2023).
O. Careta, A. Nicolenco, F. Perdikos, A. Blanquer, E. Ibañez, E. Pellicer, C. Stefani, B. Sepúlveda, J. Nogués, J. Sort, C. Nogués, Enhanced Proliferation and Differentiation of Human Osteoblasts by Remotely Controlled Magnetic-Field-Induced Electric Stimulation Using Flexible Substrates, ACS Applied Materials & Interfaces 15, 58054 (2023).
W. Cheng, X. Wang, Z. Xiong, J. Liu, Z. Liu, Y. Jin, H. Yao, T.-S. Wong, J. Ho, B. Tee, Frictionless multiphasic interface for near-ideal aero-elastic pressure sensing, Nature materials 22, 1352 (2023).
H. Fung, Y. Hayashi, V. Salo, A. Babenko, I. Zagoriy, A. Brunner, J. Ellenberg, C. Müller, S. Cuylen-Haering, J. Mahamid, Genetically encoded multimeric tags for subcellular protein localization in cryo-EM, Nature methods 20, 1900 (2023).
M. Hisham, A. Salih, H. Butt, 3D Printing of Multimaterial Contact Lenses, ACS Biomaterials Science & Engineering 9, 4381 (2023).
F. Höglsperger, B. Vos, A. Hofemeier, M. Seyfried, B. Stövesand, A. Alavizargar, L. Topp, A. Heuer, T. Betz, B. Ravoo, Rapid and reversible optical switching of cell membrane area by an amphiphilic azobenzene, Nature communications 14, 3760 (2023).
S. Jadavi, S. Dante, L. Civiero, M. Sandre, L. Bubacco, L. Tosatto, P. Bianchini, C. Canale, A. Diaspro, Fluorescence labeling methods influence the aggregation process of α-syn in vitro differently, Nanoscale 15, 8270 (2023).
Z. Kechagia, P. Sáez, M. Gómez-González, B. Canales, S. Viswanadha, M. Zamarbide, I. Andreu, T. Koorman, A. Beedle, A. Elosegui-Artola, P. Derksen, X. Trepat, M. Arroyo, P. Roca-Cusachs, The laminin-keratin link shields the nucleus from mechanical deformation and signalling, Nature materials 22, 1409 (2023).
A. Klimas, B. Gallagher, P. Wijesekara, S. Fekir, E. DiBernardo, Z. Cheng, D. Stolz, F. Cambi, S. Watkins, S. Brody, A. Horani, A. Barth, C. Moore, X. Ren, Y. Zhao, Magnify is a universal molecular anchoring strategy for expansion microscopy, Nature biotechnology 41, 858 (2023).
R. Li, M. Ebbesen, Z. Glover, T. Jæger, T. Rovers, B. Svensson, J. Brewer, A. Simonsen, R. Ipsen, A. Hougaard, Discriminating between different proteins in the microstructure of acidified milk gels by super-resolution microscopy, Food Hydrocolloids 138, 108468 (2023).
J. Liu, G. Xie, S. Lv, Q. Xiong, H. Xu, Recent applications of rolling circle amplification in biosensors and DNA nanotechnology, TrAC Trends in Analytical Chemistry 160, 116953 (2023).
Y. Ma, O. Fenton, A Unified Strategy to Improve Lipid Nanoparticle Mediated mRNA Delivery Using Adenosine Triphosphate, Journal of the American Chemical Society 145, 19800 (2023).
H. Maus, G. Hinze, S. Hammerschmidt, T. Basché, T. Schirmeister, A competition smFRET assay to study ligand-induced conformational changes of the dengue virus protease, Protein Science 32, e4526 (2023).
S. Nagella, S. Takatori, Colloidal transport phenomena in dynamic, pulsating porous materials, AIChE Journal 69, e18215 (2023).
H. Ozadam, T. Tonn, C. Han, A. Segura, I. Hoskins, S. Rao, V. Ghatpande, D. Tran, D. Catoe, M. Salit, C. Cenik, Single-cell quantification of ribosome occupancy in early mouse development, Nature 618, 1057 (2023).
J. Paris, R. Gaspar, F. Coelho, P. de Beule, B. Silva, Stability Criterion for the Assembly of Core–Shell Lipid–Polymer–Nucleic Acid Nanoparticles, ACS Nano 17, 17587 (2023).
A. Safieddine, E. Coleno, F. Lionneton, A.-M. Traboulsi, S. Salloum, C.-H. Lecellier, T. Gostan, V. Georget, C. Hassen-Khodja, A. Imbert, F. Mueller, T. Walter, M. Peter, E. Bertrand, HT-smFISH, Nature Protocols 18, 157 (2023).
W. de Santana, A. Surur, V. Momesso, P. Lopes, C. Santilli, C. Fontana, Nanocarriers for photodynamic-gene therapy, Photodiagnosis and Photodynamic Therapy 43, 103644 (2023).
C. Valentino, B. Vigani, G. Zucca, M. Ruggeri, G. Marrubini, C. Boselli, A. Icaro Cornaglia, G. Sandri, S. Rossi, Design of Novel Mechanically Resistant and Biodegradable Multichannel Platforms for the Treatment of Peripheral Nerve Injuries, Biomacromolecules (2023).
J. Waeterschoot, W. Gosselé, H. Alizadeh Zeinabad, J. Lammertyn, E. Koos, X. Casadevall i Solvas, Formation of Giant Unilamellar Vesicles Assisted by Fluorinated Nanoparticles, Environmental Toxicology 10, 2302461 (2023).
H. Wang, C. Zhu, Di Li, Visualizing enzyme catalytic process using single-molecule techniques, TrAC Trends in Analytical Chemistry 163, 117083 (2023).
G. Wen, M. Lycas, Y. Jia, V. Leen, M. Sauer, J. Hofkens, Trifunctional Linkers Enable Improved Visualization of Actin by Expansion Microscopy, ACS Nano 17, 20589 (2023).
F. Yuan, C. Lee, A. Sangani, J. Houser, L. Wang, E. Lafer, P. Rangamani, J. Stachowiak, The ins and outs of membrane bending by intrinsically disordered proteins, Science advances 9, eadg3485 (2023).
J. Zamel, J. Chen, S. Zaer, P. Harris, P. Drori, M. Lebendiker, N. Kalisman, N. Dokholyan, E. Lerner, Structural and dynamic insights into α-synuclein dimer conformations, Structure 31, 411-423.e6 (2023).
M. Zhang, W. Zhang, Y. Wei, X. Duan, M. Li, X. Hu, Y. Ma, Y.-H. Zhang, Cytosolic delivery of cell-impermeable fluorescent probes by mixtures of cell-penetrating peptides for multicolor long-term live-cell nanoscopy, Cell Reports Physical Science 4, 101674 (2023).
A. Ali, Y. Bagheri, Q. Tian, M. You, Advanced DNA Zipper Probes for Detecting Cell Membrane Lipid Domains, Nano letters 22, 7579 (2022).
V.-V. Auvinen, P. Laurén, B. Shen, J. Isokuortti, N. Durandin, T. Lajunen, V. Linko, T. Laaksonen, Nanoparticle release from anionic nanocellulose hydrogel matrix, Cellulose 29, 9707 (2022).
Y. Bagheri, A. Ali, P. Keshri, J. Chambers, A. Gershenson, M. You, Imaging Membrane Order and Dynamic Interactions in Living Cells with a DNA Zipper Probe, Angewandte Chemie International Edition 61, e202112033 (2022).
W. Chen, X.-Y. Zhang, D. Su, Z.-J. Gao, T. Zhang, Plasmonic Nanostructure-Loaded filtering film for Color Blindness Management, SID Symposium Digest of Technical Papers 53, 104 (2022).
A. Clancy, D. Chen, J. Bruns, J. Nadella, S. Stealey, Y. Zhang, A. Timperman, S. Zustiak, Hydrogel-based microfluidic device with multiplexed 3D in vitro cell culture, Scientific Reports 12, 1 (2022).
J. Danial, Y. Quintana, U. Ros, R. Shalaby, E. Margheritis, S. Chumpen Ramirez, C. Ungermann, A. Garcia-Saez, K. Cosentino, Systematic Assessment of the Accuracy of Subunit Counting in Biomolecular Complexes Using Automated Single-Molecule Brightness Analysis, The Journal of Physical Chemistry Letters 13, 822 (2022).
T. Gervais, Y. Temiz, L. Aubé, E. Delamarche, Large-Scale Dried Reagent Reconstitution and Diffusion Control Using Microfluidic Self-Coalescence Modules, Small 18, 2105939 (2022).
S. Groeer, M. Garni, A. Samanta, A. Walther, Insertion of 3D DNA Origami Nanopores into Block Copolymer Vesicles, ChemSystemsChem 4, e202200009 (2022).
J. Houser, H. Cho, C. Hayden, N. Yang, L. Wang, E. Lafer, D. Thirumalai, J. Stachowiak, Molecular mechanisms of steric pressure generation and membrane remodeling by disordered proteins, Biophysical Journal 121, 3320 (2022).
M. Iriarte-Alonso, A. Bittner, S. Chiantia, Influenza A virus hemagglutinin prevents extensive membrane damage upon dehydration, BBA Advances 2, 100048 (2022).
S. Ito, K. Hiratsuka, S. Takei, H. Nishi, D. Kitagawa, S. Kobatake, H. Miyasaka, Spatial distribution of single guest molecules along thickness of thin films of poly(2-hydroxyethyl acrylate), Photochemical & Photobiological Sciences 21, 175 (2022).
A. Jagiełło, U. Castillo, E. Botvinick, Cell mediated remodeling of stiffness matched collagen and fibrin scaffolds, Scientific Reports 12, 11736 (2022).
Z. James Glover, S. Gregersen, L. Wiking, M. Hammershøj, A. Simonsen, Microstructural changes in acid milk gels due to temperature-controlled high-intensity ultrasound treatment, International Journal of Dairy Technology 75, 321 (2022).
A. Jang, P. Praveen Kumar, D.-K. Lim, Attomolar Sensitive Magnetic Microparticles and a Surface-Enhanced Raman Scattering-Based Assay for Detecting SARS-CoV-2 Nucleic Acid Targets, ACS Applied Materials & Interfaces 14, 138 (2022).
L. Kai, Sonal, T. Heermann, P. Schwille, Reconstitution of a Reversible Membrane Switch via Prenylation by One-Pot Cell-Free Expression, ACS synthetic biology 12, 108 (2023).
K. Kamagata, N. Iwaki, S. Kanbayashi, T. Banerjee, R. Chiba, V. Gaudon, B. Castaing, S. Sakomoto, Structure-dependent recruitment and diffusion of guest proteins in liquid droplets of FUS, Scientific Reports 12, 1 (2022).
C. Kimna, M. Bauer, T. Lutz, S. Mansi, E. Akyuz, Z. Doganyigit, P. Karakol, P. Mela, O. Lieleg, Multifunctional “Janus-Type” Bilayer Films Combine Broad-Range Tissue Adhesion with Guided Drug Release, Advanced Functional Materials 32, 2105721 (2022).
M. Klučáková, M. Havlíková, F. Mravec, M. Pekař, Diffusion of dyes in polyelectrolyte-surfactant hydrogels, RSC advances 12, 13242 (2022).
A. Lebeau, D. Bruyere, P. Roncarati, P. Peixoto, E. Hervouet, G. Cobraiville, B. Taminiau, M. Masson, C. Gallego, G. Mazzucchelli, N. Smargiasso, M. Fleron, D. Baiwir, E. Hendrick, C. Pilard, T. Lerho, C. Reynders, M. Ancion, R. Greimers, J.-C. Twizere, G. Daube, G. Schlecht-Louf, F. Bachelerie, J.-D. Combes, P. Melin, M. Fillet, P. Delvenne, P. Hubert, M. Herfs, HPV infection alters vaginal microbiome through down-regulating host mucosal innate peptides used by Lactobacilli as amino acid sources, Nature Communications 13, 1 (2022).
M. Marczynski, T. Lutz, R. Schlatterer, M. Henkel, B. Balzer, O. Lieleg, Contamination with Black Carbon Nanoparticles Alters the Selective Permeability of Mucin Hydrogels, ACS Applied Nano Materials 5, 16955 (2022).
H. Maus, G. Hinze, S. Hammerschmidt, T. Basché, T. Schirmeister, A competition smFRET assay to study ligand-induced conformational changes of the dengue virus protease, Protein Science 32, e4526 (2023).
J. Poisson, Z. Hudson, Luminescent Surface-Tethered Polymer Brush Materials, Chemistry – A European Journal 28, e202200552 (2022).
U. Pramanik, A. Nandy, L. Khamari, S. Mukherjee, Structure and Transition Dynamics of Intrinsically Disordered Proteins Probed by Single-Molecule Spectroscopy, Langmuir: the ACS journal of surfaces and colloids 38, 12764 (2022).
S. Puza, S. Caesar, C. Poojari, M. Jung, R. Seemann, J. Hub, B. Schrul, J.-B. Fleury, Lipid Droplets Embedded in a Model Cell Membrane Create a Phospholipid Diffusion Barrier, Small 18, 2106524 (2022).
N. Roostaei, S. Hamidi, Plasmonic Eyeglasses Based on Gold Nanoparticles for Color Vision Deficiency Management, ACS Applied Nano Materials 5, 18788 (2022).
A. Safieddine, E. Coleno, F. Lionneton, A.-M. Traboulsi, S. Salloum, C.-H. Lecellier, T. Gostan, V. Georget, C. Hassen-Khodja, A. Imbert, F. Mueller, T. Walter, M. Peter, E. Bertrand, HT-smFISH - A cost-effective and flexible workflow for high-throughput single-molecule RNA imaging, Nature Protocols 18, 157 (2023).
I. Seitz, H. Ijäs, V. Linko, M. Kostiainen, Optically Responsive Protein Coating of DNA Origami for Triggered Antigen Targeting, ACS applied materials & interfaces 14, 38515 (2022).
Y. Shin, N. Fomina, C. Johnson, T. Rocznik, H. Ahmad, R. Staley, J. Weller, C. Lang, Toward Rapid and Automated Immunoassays, Analytical Chemistry 94, 13171 (2022).
A. Silva-Santos, P. Paulo, D. F. Prazeres, Scalable purification of single stranded DNA scaffolds for biomanufacturing DNA-origami nanostructures, Separation and Purification Technology 298, 121623 (2022).
F. Torricella, L. Barbieri, V. Bazzurro, A. Diaspro, L. Banci, Protein delivery to living cells by thermal stimulation for biophysical investigation, Scientific Reports 12, 1 (2022).
T. Walther, K. Jahnke, T. Abele, K. Göpfrich, Printing and Erasing of DNA-Based Photoresists Inside Synthetic Cells, Advanced Functional Materials 32, 2200762 (2022).
B. White, P. Kumar, A. Conwell, K. Wu, J. Baskin, Lipid Expansion Microscopy, Journal of the American Chemical Society 144, 18212 (2022).
H. Yoo, C. Davis, An in Vitro Cytomimetic of In-Cell RNA Folding, ChemBioChem 23, e202200406 (2022).
W. Zhang, L. Bertinetti, E. Yavuzsoy, C. Gao, E. Schneck, P. Fratzl, Submicron-Sized In Situ Osmotic Pressure Sensors for In Vitro Applications in Biology, Advanced Healthcare Materials n/a, 2202373 (2022).
L. Baltiansky, E. Sarafian-Tamam, E. Greenwald, O. Feinerman, Dual-fluorescence imaging and automated trophallaxis detection for studying multi-nutrient regulation in superorganisms, Methods in Ecology and Evolution 12, 1441 (2021).
N. Bauer, A. Yang, X. Wang, Y. Zhou, A. Klibanski, R. Soberman, A cross–nearest neighbor/Monte Carlo algorithm for single-molecule localization microscopy defines interactions between p53, Mdm2, and MEG3, Journal of Biological Chemistry 296, 100540 (2021).
S. Cai, T. Pataillot-Meakin, A. Shibakawa, R. Ren, C. Bevan, S. Ladame, A. Ivanov, J. Edel, Single-molecule amplification-free multiplexed detection of circulating microRNA cancer biomarkers from serum, Nature Communications 12, 1 (2021).
L. Campbell, K. Pannoni, N. Savory, D. Lal, S. Farris, Protein-retention expansion microscopy for visualizing subcellular organelles in fixed brain tissue, Journal of neuroscience methods 361, 109285 (2021).
J. Deng, A. Walther, Autonomous DNA nanostructures instructed by hierarchically concatenated chemical reaction networks, Nature Communications 12, 1 (2021).
A. Dohare, S. Sudhakar, B. Brodbeck, A. Mukherjee, M. Brecht, A. Kandelbauer, E. Schäffer, H. Mayer, Anisotropic and Amphiphilic Mesoporous Core–Shell Silica Microparticles Provide Chemically Selective Environments for Simultaneous Delivery of Curcumin and Quercetin, Langmuir 37, 13460 (2021).
I. Elbalasy, P. Mollenkopf, C. Tutmarc, H. Herrmann, J. Schnauß, Keratins determine network stress responsiveness in reconstituted actin-keratin filament systems, Soft Matter (2021).
M. Elsherif, A. Salih, A. Yetisen, H. Butt, Contact Lenses for Color Vision Deficiency, Advanced Materials Technologies 6, 2000797 (2021).
A. Gómez-Varela, R. Gaspar, A. Miranda, J. Assis, R. Valverde, M. Einicker-Lamas, B. Silva, P. de Beule, Fluorescence cross-correlation spectroscopy as a valuable tool to characterize cationic liposome-DNA nanoparticle assembly, Encyclopedia of Analytical Chemistry 14, e202000200 (2021).
C. Götz, G. Hinze, A. Gellert, H. Maus, F. von Hammerstein, S. Hammerschmidt, L. Lauth, U. Hellmich, T. Schirmeister, T. Basché, Conformational Dynamics of the Dengue Virus Protease Revealed by Fluorescence Correlation and Single-Molecule FRET Studies, The Journal of Physical Chemistry B 125, 6837 (2021).
S. Gregersen, Z. Glover, L. Wiking, A. Simonsen, K. Bertelsen, B. Pedersen, K. Poulsen, U. Andersen, M. Hammershøj, Microstructure and rheology of acid milk gels and stirred yoghurts –quantification of process-induced changes by auto- and cross correlation image analysis, Food Hydrocolloids 111, 106269 (2021).
T. Hamacher, J. Berendsen, J. van Dongen, R. van der Hee, J. Cornelissen, M. Broekhuijse, L. Segerink, Virus removal from semen with a pinched flow fractionation microfluidic chip, Lab on a Chip 21, 4477 (2021).
Hendrik A. Messal, Jorge Almagro, May Zaw Thin, Antonio Tedeschi, Alessandro Ciccarelli, Laura Blackie, Kurt I. Anderson, Irene Miguel-Aliaga, Jacco van Rheenen, Axel Behrens, Antigen retrieval and clearing for whole-organ immunofluorescence by FLASH, Nature Protocols 16, 239 (2021).
B. Ignacio, T. Bakkum, K. Bonger, N. Martin, S. van Kasteren, Metabolic labeling probes for interrogation of the host-pathogen interaction, Organic & Biomolecular Chemistry (2021).
T. Irmscher, Y. Roske, I. Gayk, V. Dunsing, S. Chiantia, U. Heinemann, S. Barbirz, Pantoea stewartii WceF is a glycan biofilm-modifying enzyme with a bacteriophage tailspike-like fold, Journal of Biological Chemistry 296, 100286 (2021).
J. Lin, K. Yang, E. New, Strategies for organelle targeting of fluorescent probes, Organic & Biomolecular Chemistry 19, 9339 (2021).
V. Maffeis, A. Belluati, I. Craciun, D. Wu, S. Novak, C.-A. Schoenenberger, C. Palivan, Clustering of catalytic nanocompartments for enhancing an extracellular non-native cascade reaction, Chemical Science 12, 12274 (2021).
A. Mahapatra, N. Mandal, K. Chattopadhyay, Cholesterol in Synaptic Vesicle Membranes Regulates the Vesicle-Binding, Function, and Aggregation of α-Synuclein, The Journal of Physical Chemistry B 125, 11099 (2021).
P. Mathiassen, A. Menon, T. Pomorski, Endoplasmic reticulum phospholipid scramblase activity revealed after protein reconstitution into giant unilamellar vesicles containing a photostable lipid reporter, Scientific Reports 11, 1 (2021).
C. Meyer, C.-A. Schoenenberger, R. Wehr, D. Wu, C. Palivan, Artificial Melanogenesis by Confining Melanin/Polydopamine Production inside Polymersomes, Macromolecular Bioscience 21, 2100249 (2021).
A. Mierczynska-Vasilev, K. Bindon, R. Gawel, P. Smith, K. Vasilev, H.-J. Butt, K. Koynov, Fluorescence correlation spectroscopy to unravel the interactions between macromolecules in wine, Food Chemistry 352, 129343 (2021).
P. Mishra, C. Rao, A. Sarkar, A. Yadav, K. Kaushik, A. Jaiswal, C. Nandi, Super-Resolution Microscopy Revealed the Lysosomal Expansion During Epigallocatechin Gallate-Mediated Apoptosis, Langmuir 37, 10818 (2021).
A. Naga, A. Kaltbeitzel, W. Wong, L. Hauer, H.-J. Butt, D. Vollmer, How a water drop removes a particle from a hydrophobic surface, Soft Matter 17, 1746 (2021).
S. Qin, S. Isbaner, I. Gregor, J. Enderlein, Doubling the resolution of a confocal spinning-disk microscope using image scanning microscopy, Nature Protocols 16, 164 (2021).
M. Rzycki, D. Drabik, K. Szostak-Paluch, B. Hanus-Lorenz, S. Kraszewski, Unraveling the mechanism of octenidine and chlorhexidine on membranes, Biophysical Journal 120, 3392 (2021).
A. Schneider, M. Kithil, M. Cardoso, M. Lehmann, C. Hackenberger, Cellular uptake of large biomolecules enabled by cell-surface-reactive cell-penetrating peptide additives, Nature Chemistry 13, 530 (2021).
S. Shi, N. Quarta, H. Zhang, Z. Lu, M. Hof, R. Šachl, R. Liu, M. Hoernke, Hidden complexity in membrane permeabilization behavior of antimicrobial polycations, Physical Chemistry Chemical Physics 23, 1475 (2021).
M. Shihan, S. Novo, S. Le Marchand, Y. Wang, M. Duncan, A simple method for quantitating confocal fluorescent images, Biochemistry and Biophysics Reports 25, 100916 (2021).
J. Sindram, M. Karg, Polymer ligand binding to surface-immobilized gold nanoparticles, Soft Matter 17, 7487 (2021).
B. Smolková, T. MacCulloch, T. Rockwood, M. Liu, S. Henry, A. Frtús, M. Uzhytchak, M. Lunova, M. Hof, P. Jurkiewicz, A. Dejneka, N. Stephanopoulos, O. Lunov, Protein Corona Inhibits Endosomal Escape of Functionalized DNA Nanostructures in Living Cells, ACS Applied Materials & Interfaces 13, 46375 (2021).
Y. Tao, L. Chen, M. Pan, F. Zhu, D. Zhu, Tailored Biosensors for Drug Screening, Efficacy Assessment, and Toxicity Evaluation, ACS Sensors 6, 3146 (2021).
A. Tarczewska, M. Kolonko-Adamska, M. Zarębski, J. Dobrucki, A. Ożyhar, B. Greb-Markiewicz, The method utilized to purify the SARS-CoV-2 N protein can affect its molecular properties, International Journal of Biological Macromolecules 188, 391 (2021).
W. Zhang, L. Bertinetti, K. Blank, R. Dimova, C. Gao, E. Schneck, P. Fratzl, Spatiotemporal Measurement of Osmotic Pressures by FRET Imaging, Angewandte Chemie International Edition 60, 6488 (2021).
T. Andresen, J. Larsen, Compositional inhomogeneity of drug delivery liposomes quantified at the single liposome level, Acta Biomaterialia 118, 207 (2020).
K. Bielec, G. Bubak, T. Kalwarczyk, R. Holyst, Analysis of Brightness of a Single Fluorophore for Quantitative Characterization of Biochemical Reactions, The Journal of Physical Chemistry B 124, 1941 (2020).
K. Cervantes-Salguero, M. Freeley, J. Chávez, M. Palma, Single-molecule DNA origami aptasensors for real-time biomarker detection, Journal of Materials Chemistry B 8, 6352 (2020).
Z. Cheng, C. Shurer, S. Schmidt, V. Gupta, G. Chuang, J. Su, A. Watkins, A. Shetty, J. Spector, C.-Y. Hui, H. Reesink, M. Paszek, The surface stress of biomedical silicones is a stimulant of cellular response, Science Advances 6, eaay0076 (2020).
B. Davis, M. O’Connell, Put on Your Para-spectacles, Molecular Cell 77, 207 (2020).
J. Deng, D. Bezold, H. Jessen, A. Walther, Multiple Light Control Mechanisms in ATP-Fueled Non-equilibrium DNA Systems, Angewandte Chemie International Edition 59, 12084 (2020).
F. Geyer, M. D'Acunzi, A. Sharifi-Aghili, A. Saal, N. Gao, A. Kaltbeitzel, T.-F. Sloot, R. Berger, H.-J. Butt, D. Vollmer, When and how self-cleaning of superhydrophobic surfaces works, Science Advances 6, eaaw9727 (2020).
S. Ghosh, N. Karedla, I. Gregor, Single-molecule confinement with uniform electrodynamic nanofluidics, Lab on a Chip 20, 3249 (2020).
Z. Glover, M. Francis, A. Bisgaard, U. Andersen, L. Johansen, M. Povey, M. Holmes, J. Brewer, A. Simonsen, Dynamic moisture loss explored through quantitative super-resolution microscopy, spatial micro-viscosity and macroscopic analyses in acid milk gels, Food Hydrocolloids 101, 105501 (2020).
J. Houser, C. Hayden, D. Thirumalai, J. Stachowiak, A Förster Resonance Energy Transfer-Based Sensor of Steric Pressure on Membrane Surfaces, Journal of the American Chemical Society 142, 20796 (2020).
C. Iserman, C. Roden, M. Boerneke, R. Sealfon, G. McLaughlin, I. Jungreis, E. Fritch, Y. Hou, J. Ekena, C. Weidmann, C. Theesfeld, M. Kellis, O. Troyanskaya, R. Baric, T. Sheahan, K. Weeks, A. Gladfelter, Genomic RNA Elements Drive Phase Separation of the SARS-CoV-2 Nucleocapsid, Molecular Cell 80, 1078-1091.e6 (2020).
A. Jimenez, K. Friedl, C. Leterrier, About samples, giving examples, Methods 174, 100 (2020).
Joshua M. Brockman, Hanquan Su, Aaron T. Blanchard, Yuxin Duan, Travis Meyer, M. Edward Quach, Roxanne Glazier, Alisina Bazrafshan, Rachel L. Bender, Anna V. Kellner, Hiroaki Ogasawara, Rong Ma, Florian Schueder, Brian G. Petrich, Ralf Jungmann, Renhao Li, Alexa L. Mattheyses, Yonggang Ke, Khalid Salaita, Live-cell super-resolved PAINT imaging of piconewton cellular traction forces, Nature Methods 17, 1018 (2020).
J. Liu, T. Hebbrecht, T. Brans, E. Parthoens, S. Lippens, C. Li, H. de Keersmaecker, W. de Vos, S. de Smedt, R. Boukherroub, J. Gettemans, R. Xiong, K. Braeckmans, Long-term live-cell microscopy with labeled nanobodies delivered by laser-induced photoporation, Nano Research 13, 485 (2020).
R. Luo, K. Göpfrich, I. Platzman, J. Spatz, DNA-Based Assembly of Multi-Compartment Polymersome Networks, Encyclopedia of Analytical Chemistry 30, 2003480 (2020).
A. Mahapatra, S. Sarkar, S. Biswas, K. Chattopadhyay, Modulation of α-Synuclein Fibrillation by Ultrasmall and Biocompatible Gold Nanoclusters, ACS chemical neuroscience 11, 3442 (2020).
G. Mohanan, K. Nair, K. Nampoothiri, H. Bajaj, Engineering bio-mimicking functional vesicles with multiple compartments for quantifying molecular transport, Chemical Science 11, 4669 (2020).
M. Olsman, V. Sereti, K. Andreassen, S. Snipstad, A. van Wamel, R. Eliasen, S. Berg, A. Urquhart, T. Andresen, C. Davies, Ultrasound-mediated delivery enhances therapeutic efficacy of MMP sensitive liposomes, Journal of Controlled Release 325, 121 (2020).
D. de Pasquale, A. Marino, C. Tapeinos, C. Pucci, S. Rocchiccioli, E. Michelucci, F. Finamore, L. McDonnell, A. Scarpellini, S. Lauciello, M. Prato, A. Larrañaga, F. Drago, G. Ciofani, Homotypic targeting and drug delivery in glioblastoma cells through cell membrane-coated boron nitride nanotubes, Materials & Design 192, 108742 (2020).
M. Safdar, A. Ghazy, M. Lastusaari, M. Karppinen, Lanthanide-based inorganic–organic hybrid materials for photon-upconversion, Journal of Materials Chemistry C 8, 6946 (2020).
B. Shen, P. Piskunen, S. Nummelin, Q. Liu, M. Kostiainen, V. Linko, Advanced DNA Nanopore Technologies, ACS Applied Bio Materials 3, 5606 (2020).
M. Štefl, K. Herbst, M. Rübsam, A. Benda, M. Knop, Single-Color Fluorescence Lifetime Cross-Correlation Spectroscopy In Vivo, Biophysical Journal 119, 1359 (2020).
W. Wong, L. Hauer, A. Naga, A. Kaltbeitzel, P. Baumli, R. Berger, M. D'Acunzi, D. Vollmer, H.‐J. Butt, Adaptive Wetting of Polydimethylsiloxane, Langmuir 36, 7236 (2020).
Y. Yano, S. Hanashima, H. Tsuchikawa, T. Yasuda, J. Slotte, E. London, M. Murata, Sphingomyelins and ent-Sphingomyelins Form Homophilic Nano-Subdomains within Liquid Ordered Domains, Biophysical Journal 119, 539 (2020).
L. Zartner, M. Muthwill, I. Dinu, C.-A. Schoenenberger, C. Palivan, The rise of bio-inspired polymer compartments responding to pathology-related signals, Journal of Materials Chemistry B 8, 6252 (2020).
S. Ballweg, E. Sezgin, M. Doktorova, R. Covino, J. Reinhard, D. Wunnicke, I. Hänelt, I. Levental, G. Hummer, R. Ernst, Regulation of lipid saturation without sensing membrane fluidity, Nature Communications 11, 1 (2020).
A. Belluati, V. Mikhalevich, S. Yorulmaz Avsar, D. Daubian, I. Craciun, M. Chami, W. Meier, C. Palivan, How Do the Properties of Amphiphilic Polymer Membranes Influence the Functional Insertion of Peptide Pores?, Biomacromolecules 21, 701 (2020).
M. Kopp, M. Linsenmeier, B. Hettich, S. Prantl, S. Stavrakis, J.-C. Leroux, P. Arosio, Microfluidic Shrinking Droplet Concentrator for Analyte Detection and Phase Separation of Protein Solutions, Analytical Chemistry 92, 5803 (2020).
A. Oberhofer, E. Reithmann, P. Spieler, W. Stepp, D. Zimmermann, B. Schmid, E. Frey, Z. Ökten, Molecular underpinnings of cytoskeletal cross-talk, Proceedings of the National Academy of Sciences 117, 3944 (2020).
R. Adão, R. Campos, E. Figueiras, P. Alpuim, J. Nieder, Graphene setting the stage, 2D Materials 6, 45056 (2019).
H. Babcock, F. Huang, C. Speer, Correcting Artifacts in Single Molecule Localization Microscopy Analysis Arising from Pixel Quantum Efficiency Differences in sCMOS Cameras, Scientific Reports 9, 1 (2019).
K. Bielec, K. Sozanski, M. Seynen, Z. Dziekan, P. ten Wolde, R. Holyst, Kinetics and equilibrium constants of oligonucleotides at low concentrations. Hybridization and melting study, Physical chemistry chemical physics : PCCP 21, 10798 (2019).
S. Cai, J. Sze, A. Ivanov, J. Edel, Small molecule electro-optical binding assay using nanopores, Nature Communications 10, 1 (2019).
M. Cosentino, C. Canale, P. Bianchini, A. Diaspro, AFM-STED correlative nanoscopy reveals a dark side in fluorescence microscopy imaging, Science Advances 5, eaav8062 (2019).
C. Dolstra, T. Rinker, S. Sankhagowit, S. Deng, C. Ting, A. Dang, T. Kuhl, D. Sasaki, Mechanism of Acid-Triggered Cargo Release from Lipid Bilayer-Coated Mesoporous Silica Particles, Langmuir 35, 10276 (2019).
M. Eriksson, M. Tuominen, M. Järn, P. Claesson, V. Wallqvist, H.‐J. Butt, D. Vollmer, M. Kappl, J. Schoelkopf, P. Gane, H. Teisala, A. Swerin, Direct Observation of Gas Meniscus Formation on a Superhydrophobic Surface, ACS Nano 13, 2246 (2019).
J. Fan, Y. Sun, Y. Xia, J. Tarbell, B. Fu, Endothelial surface glycocalyx (ESG) components and ultra-structure revealed by stochastic optical reconstruction microscopy (STORM), Biorheology 56, 77 (2019).
F. Geyer, Y. Asaumi, D. Vollmer, H.‐J. Butt, Y. Nakamura, S. Fujii, Polyhedral Liquid Marbles, Advanced Functional Materials 29 (2019).
Z. Glover, C. Ersch, U. Andersen, M. Holmes, M. Povey, J. Brewer, A. Simonsen, Super-resolution microscopy and empirically validated autocorrelation image analysis discriminates microstructures of dairy derived gels, Food Hydrocolloids 90, 62 (2019).
Y. Han, M. Li, M. Zhang, X. Huang, L. Chen, X. Hao, C. Kuang, X. Liu, Y.‐H. Zhang, Cell‐permeable organic fluorescent probes for live‐cell super‐resolution imaging of actin filaments, Journal of Chemical Technology & Biotechnology 94, 2040 (2019).
I. Iachina, I. Antonescu, J. Dreier, J. Sørensen, J. Brewer, The nanoscopic molecular pathway through human skin, Biochimica et Biophysica Acta (BBA) - General Subjects 1863, 1226 (2019).
K. Möller, T. Bein, Degradable Drug Carriers, Chemistry of Materials 31, 4364 (2019).
D. Nieves, G. Hilzenrat, J. Tran, Z. Yang, H. MacRae, M. Baker, J. Gooding, K. Gaus, tagPAINT, Royal Society Open Science 6, 191268 (2019).
A. Ohmann, K. Göpfrich, H. Joshi, R. Thompson, D. Sobota, N. Ranson, A. Aksimentiev, U. Keyser, Controlling aggregation of cholesterol-modified DNA nanostructures, Nucleic Acids Research 47, 11441 (2019).
K. Okamoto, Y. Sako, Single-Molecule Förster Resonance Energy Transfer Measurement Reveals the Dynamic Partially Ordered Structure of the Epidermal Growth Factor Receptor C-Tail Domain, The Journal of Physical Chemistry B 123, 571 (2019).
N. Sarangi, J. Basu, Preferential binding and re-organization of nanoscale domains on model lipid membranes by pore-forming toxins, Journal of Physics D: Applied Physics 52, 504001 (2019).
F. Schueder, J. Stein, F. Stehr, A. Auer, B. Sperl, M. Strauss, P. Schwille, R. Jungmann, An order of magnitude faster DNA-PAINT imaging by optimized sequence design and buffer conditions, Nature Methods 16, 1101 (2019).
S. Shah, A. Dubey, J. Reif, Improved Optical Multiplexing with Temporal DNA Barcodes, ACS synthetic biology 8, 1100 (2019).
J. Sindram, K. Volk, P. Mulvaney, M. Karg, Silver Nanoparticle Gradient Arrays, Langmuir 35, 8776 (2019).
A. Trementozzi, Z. Imam, M. Mendicino, C. Hayden, J. Stachowiak, Liposome-Mediated Chemotherapeutic Delivery Is Synergistically Enhanced by Ternary Lipid Compositions and Cationic Lipids, Langmuir 35, 12532 (2019).
D. Voerman, M. Schluck, J. Weiden, B. Joosten, L. Eggermont, T. van den Eijnde, B. Ignacio, A. Cambi, C. Figdor, P. Kouwer, M. Verdoes, R. Hammink, A. Rowan, Synthetic Semiflexible and Bioactive Brushes, Biomacromolecules 20, 2587 (2019).
O. Wade, J. Woehrstein, P. Nickels, S. Strauss, F. Stehr, J. Stein, F. Schueder, M. Strauss, M. Ganji, J. Schnitzbauer, H. Grabmayr, P. Yin, P. Schwille, R. Jungmann, 124-Color Super-resolution Imaging by Engineering DNA-PAINT Blinking Kinetics, Nano letters (2019).
O. Wahlsten, F. Ulander, D. Midtvedt, M. Henningson, V. Zhdanov, B. Agnarsson, F. Höök, Quantitative Detection of Biological Nanoparticles in Solution via Their Mediation of Colocalization of Fluorescent Liposomes, Physical Review Applied 12 (2019).
O. Wrede, S. Großkopf, T. Seidel, T. Hellweg, Dynamics of proteins confined in non-ionic bicontinuous microemulsions, Physical Chemistry Chemical Physics 21, 6725 (2019).
S. Yamashiro, D. Taniguchi, S. Tanaka, T. Kiuchi, D. Vavylonis, N. Watanabe, Convection-Induced Biased Distribution of Actin Probes in Live Cells, Biophysical Journal 116, 142 (2019).
W. Zeno, A. Thatte, L. Wang, W. Snead, E. Lafer, J. Stachowiak, Molecular Mechanisms of Membrane Curvature Sensing by a Disordered Protein, Journal of the American Chemical Society 141, 10361 (2019).
M. Zhang, M. Li, W. Zhang, Y. Han, Y.-H. Zhang, Simple and efficient delivery of cell-impermeable organic fluorescent probes into live cells for live-cell superresolution imaging, Light: Science & Applications 8, 1 (2019).
D. Zhu, S. Roy, Z. Liu, H. Weller, W. Parak, N. Feliu, Remotely controlled opening of delivery vehicles and release of cargo by external triggers, Advanced Drug Delivery Reviews 138, 117 (2019).
I. Zilkowski, F. Ziouti, A. Schulze, S. Hauck, S. Schmidt, L. Mainz, M. Sauer, K. Albrecht, F. Jundt, J. Groll, Nanogels Enable Efficient miRNA Delivery and Target Gene Downregulation in Transfection-Resistant Multiple Myeloma Cells, Biomacromolecules 20, 916 (2019).
M. Boysen, R. Kityk, M. Mayer, Hsp70- and Hsp90-Mediated Regulation of the Conformation of p53 DNA Binding Domain and p53 Cancer Variants, Molecular Cell 74, 831-843.e4 (2019).
R. Jurado, J. Adamcik, M. López-Haro, J. González-Vera, Á. Ruiz-Arias, A. Sánchez-Ferrer, R. Cuesta, J. Domínguez-Vera, J. Calvino, A. Orte, R. Mezzenga, N. Gálvez, Apoferritin Protein Amyloid Fibrils with Tunable Chirality and Polymorphism, Journal of the American Chemical Society 141, 1606 (2019).
L. Schaedel, S. Triclin, D. Chrétien, A. Abrieu, C. Aumeier, J. Gaillard, L. Blanchoin, M. Théry, K. John, Lattice defects induce microtubule self-renewal, Nature Physics 47, 97 (2019).
M. Starr, R. Sparks, A. Arango, L. Hurst, Z. Zhao, M. Lihan, J. Jenkins, E. Tajkhorshid, R. Fratti, Phosphatidic acid induces conformational changes in Sec18 protomers that prevent SNARE priming, Journal of Biological Chemistry 294, 3100 (2019).
D. Wu, V. Mihali, A. Honciuc, pH-Responsive Pickering Foams Generated by Surfactant-Free Soft Hydrogel Particles, Langmuir 35, 212 (2019).
A. Becker, M. Leskau, B. Schlingmann-Molina, S. Hohmeier, S. Alnajjar, H. Escobar, A. Ngezahayo, Functionalization of gold-nanoparticles by the Clostridium perfringens enterotoxin C-terminus for tumor cell ablation using the gold nanoparticle-mediated laser perforation technique, Scientific Reports 8, 14963 (2018).
S. Braun, Š. Pokorná, R. Šachl, M. Hof, H. Heerklotz, M. Hoernke, Biomembrane Permeabilization, ACS Nano 12, 813 (2018).
R. Chelladurai, K. Debnath, N. Jana, J. Basu, Nanoscale Heterogeneities Drive Enhanced Binding and Anomalous Diffusion of Nanoparticles in Model Biomembranes, Langmuir 34, 1691 (2018).
A. Chizhik, C. Wollnik, D. Ruhlandt, N. Karedla, A. Chizhik, L. Hauke, D. Hähnel, I. Gregor, J. Enderlein, F. Rehfeldt, Dual-color metal-induced and förster resonance energy transfer for cell nanoscopy, Molecular Biology of the Cell (2018).
S. Chu, A. Brown, J. Culver, R. Ghodssi, Tobacco Mosaic Virus as a Versatile Platform for Molecular Assembly and Device Fabrication, Biotechnology Journal 13 (2018).
L. Corns, S. Johnson, T. Roberts, K. Ranatunga, A. Hendry, F. Ceriani, S. Safieddine, K. Steel, A. Forge, C. Petit, D. Furness, C. Kros, W. Marcotti, Mechanotransduction is required for establishing and maintaining mature inner hair cells and regulating efferent innervation, Nature Communications 9, 4015 (2018).
J. de Vries, S. Schnichels, J. Hurst, L. Strudel, A. Gruszka, M. Kwak, K.-U. Bartz-Schmidt, M. Spitzer, A. Herrmann, DNA nanoparticles for ophthalmic drug delivery, Biomaterials 157, 98 (2018).
M. Garni, T. Einfalt, R. Goers, C. Palivan, W. Meier, Live Follow-Up of Enzymatic Reactions Inside the Cavities of Synthetic Giant Unilamellar Vesicles Equipped with Membrane Proteins Mimicking Cell Architecture, ACS synthetic biology 7, 2116 (2018).
O. Golfetto, D. Wakefield, E. Cacao, K. Avery, V. Kenyon, R. Jorand, S. Tobin, S. Biswas, J. Gutierrez, R. Clinton, Y. Ma, D. Horne, J. Williams, T. Jovanovic-Talisman, A Platform To Enhance Quantitative Single Molecule Localization Microscopy, Journal of the American Chemical Society 140, 12785 (2018).
A. Jo, R. Zhang, I. Allen, J. Riffle, R. Davis, Design and Fabrication of Streptavidin-Functionalized, Fluorescently Labeled Polymeric Nanocarriers, Langmuir 34, 15783 (2018).
T. Litschel, K. Ganzinger, T. Movinkel, M. Heymann, T. Robinson, H. Mutschler, P. Schwille, Freeze-thaw cycles induce content exchange between cell-sized lipid vesicles, New Journal of Physics 20, 55008 (2018).
M. Roccio, M. Perny, M. Ealy, H. Widmer, S. Heller, P. Senn, Molecular characterization and prospective isolation of human fetal cochlear hair cell progenitors, Nature Communications 9, 4027 (2018).
F. Roncato, F. Rruga, E. Porcù, E. Casarin, R. Ronca, F. Maccarinelli, N. Realdon, G. Basso, R. Alon, G. Viola, M. Morpurgo, Improvement and extension of anti-EGFR targeting in breast cancer therapy by integration with the Avidin-Nucleic-Acid-Nano-Assemblies, Nature Communications 9, 4070 (2018).
F. Ruggeri, M. Krishnan, Spectrally resolved single-molecule electrometry, The Journal of Chemical Physics 148, 123307 (2018).
A. Sanchez, G. Kemper, S. Serpa, L. Melgar, P. Milon, A Smartphone-Enabled, Portable and Stand-Alone Fluorescence Quantitation System, Proceedings of the 2018 IEEE XXV International Conference on Electronics, Electrical Engineering and Computing (INTERCON), IEEE Piscataway, NJ, 2018, 1.
L. Sánchez‐García, N. Serna, P. Alamo, R. Sala, M. Virtudes Cespedes, M. Roldan, A. Sánchez‐Chardi, U. Unzueta, I. Casanova, R. Mangues, E. Vázquez, A. Villaverde, Self-assembling toxin-based nanoparticles as self-delivered antitumoral drugs, Journal of Controlled Release 274, 81 (2018).
N. Sarangi, C. Roobala, J. Basu, Unraveling complex nanoscale lipid dynamics in simple model biomembranes, Methods 140-141, 198 (2018).
C. Slagle, D. Thamm, E. Randall, M. Borden, Click Conjugation of Cloaked Peptide Ligands to Microbubbles, Bioconjugate Chemistry 29, 1534 (2018).
S. Thekkan, M. Jani, C. Cui, K. Dan, G. Zhou, L. Becker, Y. Krishnan, A DNA-based fluorescent reporter maps HOCl production in the maturing phagosome, Nature chemical biology, 1 (2018).
J. Wang, R. Deng, Energy Transfer in Dye‐Coupled Lanthanide‐Doped Nanoparticles, Chemistry – An Asian Journal 13, 614 (2018).
L. Weger, M. Weidmann, W. Ali, M. Hildebrandt, J. Gutmann, K. Hoffmann-Jacobsen, Polymer Diffusion in the Interphase Between Surface and Solution, Langmuir 34, 7021 (2018).
R. Chéreau, G. Saraceno, J. Angibaud, D. Cattaert, U. Nägerl, Superresolution imaging reveals activity-dependent plasticity of axon morphology linked to changes in action potential conduction velocity, Proceedings of the National Academy of Sciences 114, 1401 (2017).
R. Diekmann, Ø. Helle, C. Øie, P. McCourt, T. Huser, M. Schüttpelz, B. Ahluwalia, Chip-based wide field-of-view nanoscopy, Nature Photonics 11, 322 (2017).
S. Honghong, J. Feng, J. Li, Y. Xia, D. Yao, Application of CLARITY to Investigate the 3D Architecture of 14-3-3 Zeta Protein in AD, Int. J. Biomed. Sci. Eng. 5 (2017).
A. Irimie, L. Sonea, A. Jurj, N. Mehterov, A. Zimta, L. Budisan, C. Braicu, I. Berindan-Neagoe, Future trends and emerging issues for nanodelivery systems in oral and oropharyngeal cancer, International journal of nanomedicine 12, 4593 (2017).
J. Scrimgeour, L. McLane, P. Chang, J. Curtis, Single-Molecule Imaging of Proteoglycans in the Pericellular Matrix, Biophysical Journal 113, 2316 (2017).
M. Wang, H. Jin, Spray-Induced Gene Silencing, Trends in microbiology 25, 4 (2017).
X. Wang, R. Valiev, T. Ohulchanskyy, H. Ågren, C. Yang, G. Chen, Dye-sensitized lanthanide-doped upconversion nanoparticles, Chemical Society Reviews 46, 4150 (2017).
L. Weger, K. Hoffmann-Jacobsen, A total internal reflection-fluorescence correlation spectroscopy setup with pulsed diode laser excitation, Review of Scientific Instruments 88, 93102 (2017).